You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_551011-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_551011-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_551011-t26_1-p1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH140 | 35 | 500 | 3e-81 | 0.9393203883495146 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404153 | DUF4038 | 5.02e-123 | 35 | 412 | 2 | 320 | Protein of unknown function (DUF4038). A family of putative cellulases. |
| 403955 | Collagen_bind_2 | 8.74e-15 | 443 | 529 | 12 | 92 | Putative collagen-binding domain of a collagenase. This domain is likely to be the collagen-binding domain of a family of bacterial collagenase enzymes. It is the C-terminal part of the Structure 3kzs (information derived from TOPSAN). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.21e-171 | 1 | 529 | 1 | 534 | |
| 1.26e-160 | 16 | 528 | 19 | 535 | |
| 9.51e-76 | 36 | 529 | 23 | 518 | |
| 1.59e-74 | 20 | 529 | 17 | 508 | |
| 2.38e-73 | 32 | 529 | 31 | 508 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.33e-21 | 24 | 528 | 13 | 444 | Glycoside hydrolase BT_1012 [Bacteroides thetaiotaomicron VPI-5482],5MSY_B Glycoside hydrolase BT_1012 [Bacteroides thetaiotaomicron VPI-5482],5MSY_C Glycoside hydrolase BT_1012 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 2.61e-19 | 24 | 528 | 13 | 444 | Crystal structure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution [Bacteroides thetaiotaomicron VPI-5482],3KZS_B Crystal structure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution [Bacteroides thetaiotaomicron VPI-5482],3KZS_C Crystal structure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution [Bacteroides thetaiotaomicron VPI-5482],3KZS_D Crystal structure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
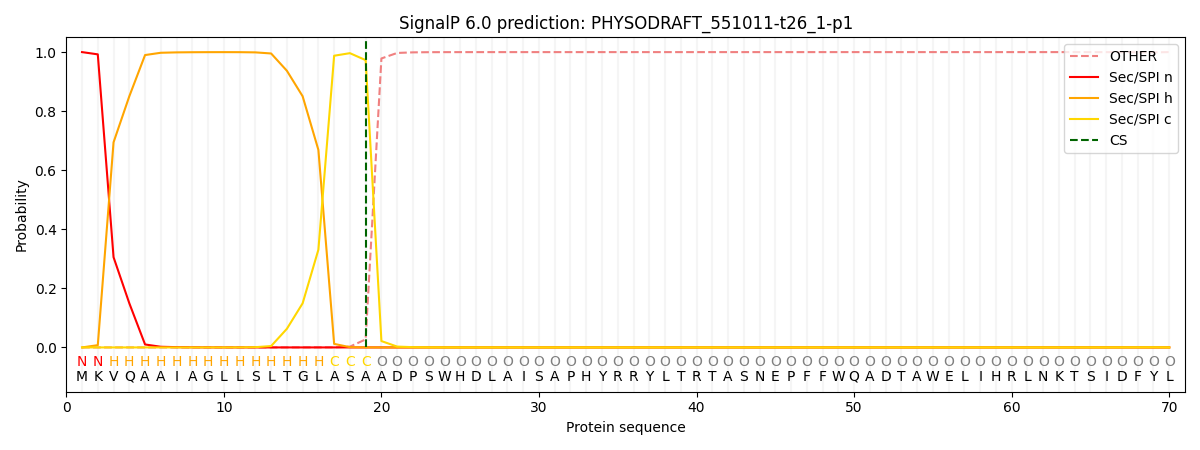
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001199 | 0.998782 | CS pos: 19-20. Pr: 0.9728 |
