You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_497435-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_497435-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
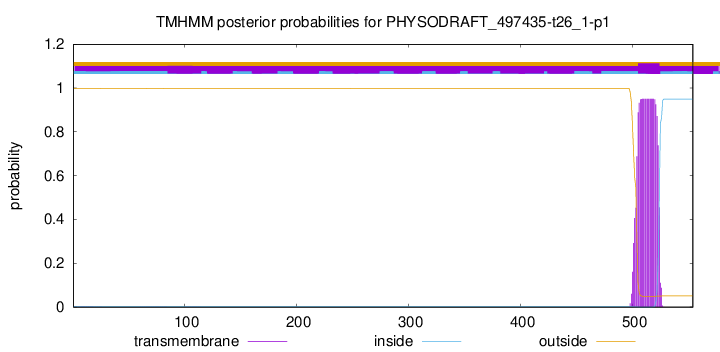
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_497435-t26_1-p1 | |||||||||||
| CAZy Family | GH81 | |||||||||||
| CAZyme Description | glycoside hydrolase family 17-like protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 23 | 278 | 2.1e-18 | 0.9903536977491961 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.07e-23 | 12 | 270 | 33 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 411384 | Streccoc_I_II | 2.20e-07 | 296 | 405 | 871 | 988 | antigen I/II family LPXTG-anchored adhesin. Members of the antigen I/II family are adhesins with a glucan-binding domain, two types of repetitive regions, an isopeptide bond-forming domain associated with shear resistance, and a C-terminal LPXTG motif for anchoring to the cell wall. They occur in oral Streptococci, and tend to be major cell surface adhesins. Members of this family include SspA and SspB from Streptococcus gordonii, antigen I/II from S. mutans, etc. |
| 236090 | PRK07764 | 3.93e-07 | 266 | 432 | 569 | 735 | DNA polymerase III subunits gamma and tau; Validated |
| 411474 | fibronec_FbpA | 4.46e-07 | 296 | 432 | 167 | 292 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
| 411474 | fibronec_FbpA | 1.46e-06 | 278 | 434 | 135 | 275 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.27e-118 | 1 | 274 | 1 | 274 | |
| 6.27e-24 | 6 | 270 | 6 | 280 | |
| 3.44e-23 | 23 | 274 | 94 | 374 | |
| 2.10e-22 | 16 | 274 | 567 | 829 | |
| 2.54e-22 | 23 | 276 | 23 | 271 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.36e-10 | 23 | 280 | 39 | 295 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.28e-12 | 23 | 252 | 297 | 530 | Probable beta-glucosidase btgE OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgE PE=3 SV=1 |
|
| 1.25e-11 | 23 | 252 | 310 | 543 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
|
| 1.25e-11 | 23 | 252 | 310 | 543 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 3.78e-11 | 23 | 220 | 304 | 506 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
|
| 9.37e-10 | 21 | 220 | 136 | 337 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000228 | 0.999729 | CS pos: 21-22. Pr: 0.9797 |