You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_492444-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_492444-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_492444-t26_1-p1 | |||||||||||
| CAZy Family | PL3|PL3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.170:2 |
|---|
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 17 | 720 | 15 | 721 | |
| 2.78e-15 | 63 | 382 | 75 | 348 | |
| 2.89e-15 | 63 | 382 | 25 | 305 | |
| 9.08e-15 | 63 | 382 | 25 | 305 | |
| 1.51e-14 | 63 | 338 | 58 | 300 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.37e-09 | 63 | 335 | 45 | 272 | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with serine and glycerol [Mycolicibacterium hassiacum DSM 44199],5OI1_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with serine and glycerol [Mycolicibacterium hassiacum DSM 44199],5OIW_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with glucosylglycerate [Mycolicibacterium hassiacum DSM 44199],5OIW_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with glucosylglycerate [Mycolicibacterium hassiacum DSM 44199],5OJ4_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with mannosylglycerate [Mycolicibacterium hassiacum DSM 44199],5OJ4_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with mannosylglycerate [Mycolicibacterium hassiacum DSM 44199],5ONZ_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with glucosylglycolate [Mycolicibacterium hassiacum DSM 44199],5ONZ_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D182A variant in complex with glucosylglycolate [Mycolicibacterium hassiacum DSM 44199] |
|
| 9.74e-09 | 63 | 335 | 45 | 272 | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) in complex with glycerol [Mycolicibacterium hassiacum DSM 44199],5OHC_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) in complex with glycerol [Mycolicibacterium hassiacum DSM 44199],5OI0_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) in complex with serine and glycerol [Mycolicibacterium hassiacum DSM 44199],5OI0_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) in complex with serine and glycerol [Mycolicibacterium hassiacum DSM 44199],6Q5T_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) - apo form [Mycolicibacterium hassiacum DSM 44199],6Q5T_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) - apo form [Mycolicibacterium hassiacum DSM 44199] |
|
| 9.74e-09 | 63 | 335 | 45 | 272 | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with serine and glycerol [Mycolicibacterium hassiacum DSM 44199],5OIE_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with serine and glycerol [Mycolicibacterium hassiacum DSM 44199],5OJU_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with glucosylglycerate [Mycolicibacterium hassiacum DSM 44199],5OJU_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with glucosylglycerate [Mycolicibacterium hassiacum DSM 44199],5OJV_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with mannosylglycerate [Mycolicibacterium hassiacum DSM 44199],5OJV_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with mannosylglycerate [Mycolicibacterium hassiacum DSM 44199],5ONT_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase(MhGgH) E419A variant in complex with glucosylglycerol [Mycolicibacterium hassiacum DSM 44199],5ONT_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase(MhGgH) E419A variant in complex with glucosylglycerol [Mycolicibacterium hassiacum DSM 44199],5OO2_A Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with glucosylglycolate [Mycolicibacterium hassiacum DSM 44199],5OO2_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with glucosylglycolate [Mycolicibacterium hassiacum DSM 44199] |
|
| 9.74e-09 | 63 | 335 | 45 | 272 | Mycobacterial hydrolase complex 14. [Mycolicibacterium hassiacum DSM 44199],6G3N_B Mycobacterial hydrolase complex 14. [Mycolicibacterium hassiacum DSM 44199] |
|
| 5.19e-08 | 64 | 335 | 46 | 272 | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D43A variant in complex with serine and glycerol [Mycolicibacterium hassiacum DSM 44199],5OIV_B Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) D43A variant in complex with serine and glycerol [Mycolicibacterium hassiacum DSM 44199] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.99e-08 | 63 | 335 | 43 | 270 | Glucosylglycerate hydrolase OS=Mycolicibacterium hassiacum (strain DSM 44199 / CIP 105218 / JCM 12690 / 3849) OX=1122247 GN=ggh PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
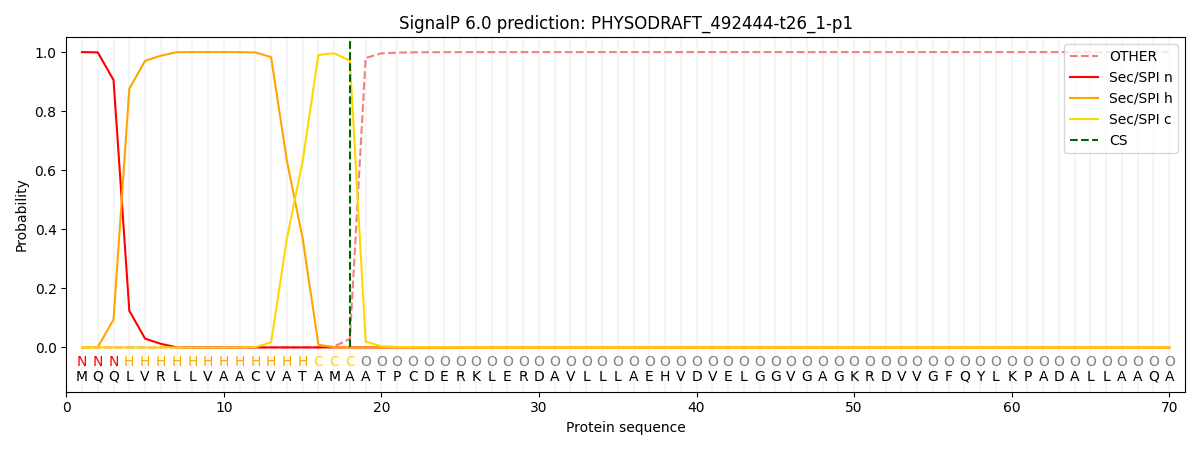
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000299 | 0.999677 | CS pos: 18-19. Pr: 0.9709 |

