You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_477860-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_477860-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_477860-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.10.3.2:4 | 1.10.3.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 67 | 539 | 2e-32 | 0.9692737430167597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 259922 | CuRO_1_Tth-MCO_like | 1.48e-51 | 46 | 170 | 1 | 138 | The first cupredoxin domain of the bacterial laccases similar to Tth-MCO from Thermus Thermophilus. The subfamily of bacterial laccases includes Tth-MCO and similar proteins. Tth-MCO is a hyperthermophilic multicopper oxidase (MCO) from thermus thermophilus HB27. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism in fungi and plants. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 1 of 3-domain MCOs contains part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 225043 | SufI | 3.27e-37 | 72 | 544 | 59 | 448 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| 259869 | CuRO_1_LCC_like | 7.95e-26 | 50 | 170 | 4 | 119 | Cupredoxin domain 1 of laccase-like multicopper oxidases; including laccase, CueO, spore coat protein A, ascorbate oxidase and similar proteins. Laccase-like multicopper oxidases (MCOs) in this family contain three cupredoxin domains. They are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. Although the members of this family have diverse functions, majority of them have three cupredoxin domain repeats. The copper ions are bound in several sites; Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 1 of 3-domain MCOs contains part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. Also included in this family are cupredoxin domains 1, 3, and 5 of the 6-domain MCO ceruloplasmin and similar proteins. |
| 400195 | Cu-oxidase_3 | 5.96e-25 | 70 | 174 | 20 | 118 | Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognized by the pfam00394 model. |
| 259930 | CuRO_1_CumA_like | 3.39e-19 | 70 | 170 | 25 | 118 | The first cupredoxin domain of CumA like multicopper oxidase. This multicopper oxidase (MCO) subfamily includes CumA from Pseudomonas putida, which is involved in the oxidation of Mn(II). However, the cumA gene has been identified in a variety of bacterial species, including both Mn(II)-oxidizing and non-Mn(II)-oxidizing strains. Thus, the proteins in this family may catalyze the oxidation of other substrates. MCO catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water and has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 1 of 3-domain MCOs contains part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.21e-46 | 101 | 544 | 5 | 468 | |
| 1.13e-31 | 66 | 548 | 9 | 479 | |
| 5.44e-10 | 47 | 334 | 188 | 457 | |
| 7.93e-10 | 73 | 539 | 96 | 562 | |
| 3.37e-09 | 43 | 544 | 29 | 548 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.52e-27 | 69 | 544 | 74 | 479 | Multicopper oxidase from Campylobacter jejuni: a metallo-oxidase [Campylobacter jejuni subsp. jejuni] |
|
| 9.34e-22 | 69 | 544 | 39 | 438 | Crystal structure of Laccase from Thermus thermophilus HB27 [Thermus thermophilus HB27],2XUW_A Crystal Structure of Apolaccase from Thermus thermophilus HB27 [Thermus thermophilus HB27],2XVB_A Crystal structure of Laccase from Thermus thermophilus HB27 complexed with Hg, crystal of the apoenzyme soaked for 5 min. in 5 mM HgCl2 at 278 K. [Thermus thermophilus HB27],2YAE_A X-ray induced reduction of laccase from Thermus thermophilus HB27(0.0- 12.5 percent dose) [Thermus thermophilus HB27],2YAF_A X-ray induced reduction of laccase from Thermus thermophilus HB27 (12. 5-25.0 percent dose) [Thermus thermophilus HB27],2YAH_A X-ray induced reduction of laccase from Thermus thermophilus HB27 (25. 0-37.5 percent dose) [Thermus thermophilus HB27],2YAM_A X-ray induced reduction of laccase from Thermus thermophilus HB27 (37. 5-50.0 percent dose) [Thermus thermophilus HB27],2YAO_A X-ray induced reduction of laccase from Thermus thermophilus HB27 (50. 0-62.5 percent dose) [Thermus thermophilus HB27],2YAP_A X-ray induced reduction of laccase from Thermus thermophilus HB27 (62. 5-75.0 percent dose) [Thermus thermophilus HB27],2YAQ_A X-ray induced reduction of laccase from Thermus thermophilus HB27 (75. 0-87.5 percent dose) [Thermus thermophilus HB27],2YAR_A X-ray induced reduction of laccase from Thermus thermophilus HB27 (87. 5-100.0 percent dose) [Thermus thermophilus HB27],4AI7_A Crystal structure of Laccase from Thermus thermophilus HB27 complexed with Hg, crystal of the apoenzyme soaked for 2 h in 5 mM HgCl2 at 278 K. [Thermus thermophilus HB27],5AFA_A Crystal structure of Laccase from Thermus thermophilus HB27 complexed with Ag, crystal of the holoenzyme soaked for 30 m in 5 mM AgNO3 at 278 K. [Thermus thermophilus HB27],5G3B_A Preserving metallic sites affected by radiation damage: the CuT2 case in Thermus thermophilus multicopper oxidase [Thermus thermophilus],5G3C_A Preserving Metallic sites affected by radiation damage the CuT2 case in thermus termophilus multicopper oxidase [Thermus thermophilus],5G3D_A preserving Metallic Sites Affected by Radiation Damage the CuT2 cCase in Thermus Thermophilus Multicopper Oxidase [Thermus thermophilus],5G3E_A Preserving Metallic Sites Affected by Radiation DAmage the CuT2 CAse in THermus Thermophilus Multicopper Oxidase [Thermus thermophilus],5G3F_A Preserving Metallic Sites Affected by Radiation Damage the CuT2 CAse in Thermus Thermophilus Multicopper Oxidase [Thermus thermophilus],5G3G_A Preserving MEtallic Sites Affected by Radiation Damage the CuT2 case in Thermus Thermophilus multicopper Oxidase [Thermus thermophilus],5G3H_A Preserving Metallic Sites Affected by Radiation Damage the CuT2 Case in Thermus Thermophilus Multicopper oxidase [Thermus thermophilus],5JRR_A Crystal structure of native laccase from Thermus thermophilus HB27 [Thermus thermophilus HB27],5JX9_A Crystal structure of laccase from Thermus thermophilus HB27 (Cu(II)-cyclophanes, 5 min) [Thermus thermophilus HB27],5K0D_A Crystal structure of laccase from Thermus thermophilus HB27 (Cu(II)-cyclophanes, 3 min) [Thermus thermophilus HB27],5K15_A Crystal structure of laccase from Thermus thermophilus HB27 (Cu2PO, 8 min) [Thermus thermophilus HB27],5K3K_A Crystal structure of laccase from Thermus thermophilus HB27 (CuSO4, 20 min) [Thermus thermophilus HB27],5K5K_A Crystal structure of laccasse from Thermus thermophilus HB27 (ascorbic acid 10 min) [Thermus thermophilus HB27],5K7A_A Crystal structure of laccase fron Thermus thermophilus HB27 (sodium nitrate 1.5 min) [Thermus thermophilus HB27],5K84_A Crystal structure of laccase from Thermus thermophilus HB27 (sodium nitrate 10 min) [Thermus thermophilus HB27],6Q29_A Chain A, Laccase [Thermus thermophilus HB27],6TYR_A Chain A, Laccase [Thermus thermophilus HB27] |
|
| 9.42e-22 | 69 | 544 | 40 | 439 | Chain A, Laccase [Thermus thermophilus HB27],6W2K_B Chain B, Laccase [Thermus thermophilus HB27],6W9X_A Chain A, Laccase [Thermus thermophilus HB27],6WCG_A Chain A, Laccase [Thermus thermophilus HB27],6WCH_A Chain A, Laccase [Thermus thermophilus HB27],6WCL_A Chain A, Laccase [Thermus thermophilus HB27],6WCM_A Chain A, Laccase [Thermus thermophilus HB27] |
|
| 1.13e-21 | 69 | 544 | 62 | 461 | Chain A, Laccase [Thermus thermophilus HB27],6WCP_A Chain A, Laccase [Thermus thermophilus HB27] |
|
| 4.69e-16 | 64 | 333 | 21 | 240 | Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_B Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_C Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_D Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_E Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_F Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.41e-23 | 66 | 544 | 71 | 445 | Multicopper oxidase mco OS=Staphylococcus aureus OX=1280 GN=mco PE=1 SV=2 |
|
| 8.60e-23 | 66 | 544 | 71 | 445 | Multicopper oxidase mco OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=mco PE=3 SV=2 |
|
| 1.15e-22 | 66 | 546 | 71 | 447 | Multicopper oxidase mco OS=Staphylococcus haemolyticus (strain JCSC1435) OX=279808 GN=mco PE=3 SV=2 |
|
| 8.94e-22 | 66 | 546 | 71 | 447 | Multicopper oxidase mco OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=mco PE=3 SV=2 |
|
| 1.17e-09 | 51 | 336 | 69 | 326 | Multicopper oxidase MmcO OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=mmcO PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
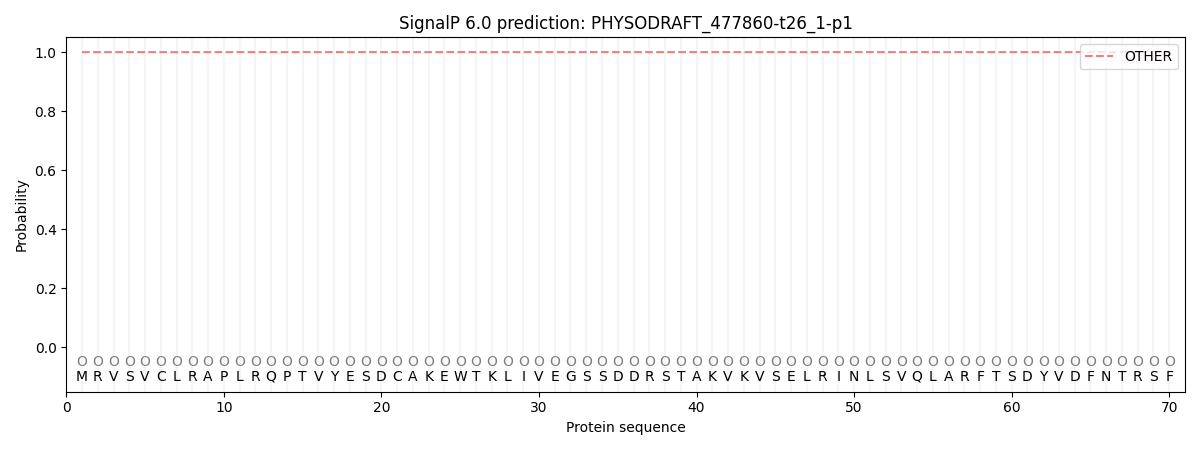
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000047 | 0.000004 |
