You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_469873-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_469873-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_469873-t26_1-p1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.260:8 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 12 | 402 | 3.8e-80 | 0.9151670951156813 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 4.44e-37 | 22 | 426 | 11 | 412 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.39e-100 | 4 | 441 | 6 | 423 | |
| 2.15e-99 | 21 | 500 | 12 | 478 | |
| 1.27e-98 | 21 | 499 | 40 | 505 | |
| 3.53e-98 | 21 | 500 | 12 | 478 | |
| 5.06e-98 | 21 | 499 | 12 | 477 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.29e-98 | 7 | 480 | 8 | 462 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Homo sapiens OX=9606 GN=ALG12 PE=1 SV=1 |
|
| 6.72e-97 | 13 | 480 | 17 | 460 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Mus musculus OX=10090 GN=Alg12 PE=2 SV=2 |
|
| 8.72e-93 | 11 | 505 | 2 | 484 | Probable Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Drosophila melanogaster OX=7227 GN=Alg12 PE=2 SV=1 |
|
| 6.48e-90 | 11 | 463 | 17 | 447 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Arabidopsis thaliana OX=3702 GN=ALG12 PE=1 SV=1 |
|
| 5.76e-86 | 11 | 467 | 5 | 443 | Probable Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Caenorhabditis elegans OX=6239 GN=algn-12 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
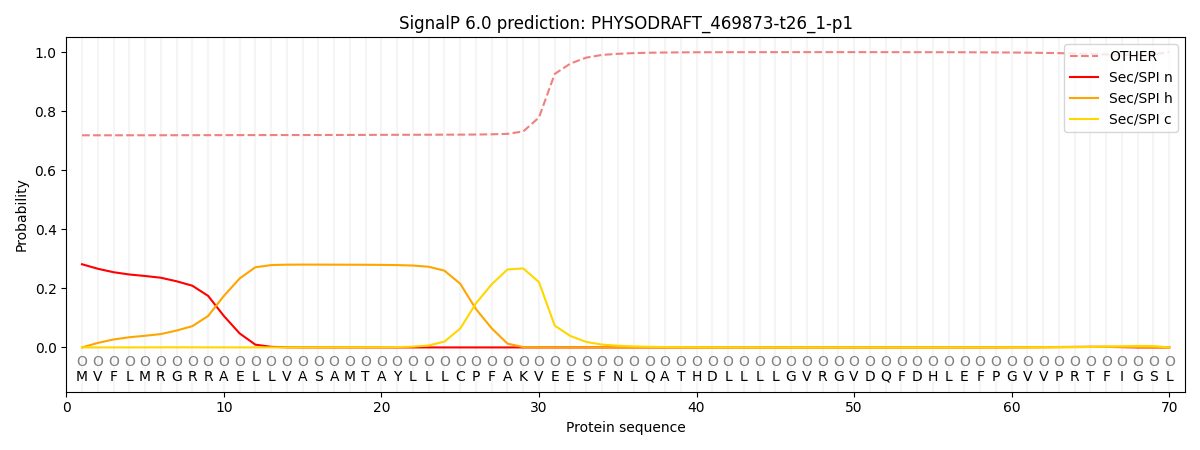
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.733451 | 0.266559 |
TMHMM Annotations download full data without filtering help

| Start | End |
|---|---|
| 99 | 121 |
| 141 | 163 |
| 170 | 192 |
| 202 | 223 |
| 267 | 289 |
| 309 | 328 |
| 335 | 352 |
| 362 | 384 |
