You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_441237-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_441237-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_441237-t26_1-p1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT50 | 78 | 272 | 1.4e-64 | 0.7099236641221374 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178434 | PLN02841 | 7.92e-85 | 1 | 272 | 52 | 340 | GPI mannosyltransferase |
| 252941 | Mannosyl_trans | 2.05e-48 | 78 | 272 | 1 | 184 | Mannosyltransferase (PIG-M). PIG-M has a DXD motif. The DXD motif is found in many glycosyltransferases that utilize nucleotide sugars. It is thought that the motif is involved in the binding of a manganese ion that is required for association of the enzymes with nucleotide sugar substrates. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.95e-91 | 5 | 272 | 54 | 318 | |
| 5.68e-72 | 1 | 272 | 62 | 334 | |
| 5.68e-72 | 1 | 272 | 62 | 334 | |
| 5.68e-72 | 1 | 272 | 62 | 334 | |
| 5.68e-72 | 1 | 272 | 62 | 334 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.01e-72 | 1 | 272 | 62 | 334 | GPI mannosyltransferase 1 OS=Homo sapiens OX=9606 GN=PIGM PE=1 SV=1 |
|
| 1.01e-72 | 1 | 272 | 62 | 334 | GPI mannosyltransferase 1 OS=Pongo abelii OX=9601 GN=PIGM PE=2 SV=1 |
|
| 3.60e-72 | 1 | 272 | 58 | 332 | GPI mannosyltransferase 1 OS=Xenopus tropicalis OX=8364 GN=pigm PE=2 SV=1 |
|
| 8.90e-71 | 1 | 272 | 62 | 334 | GPI mannosyltransferase 1 OS=Macaca fascicularis OX=9541 GN=PIGM PE=2 SV=1 |
|
| 9.91e-70 | 1 | 272 | 62 | 334 | GPI mannosyltransferase 1 OS=Bos taurus OX=9913 GN=PIGM PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000034 | 0.000003 |
TMHMM Annotations download full data without filtering help
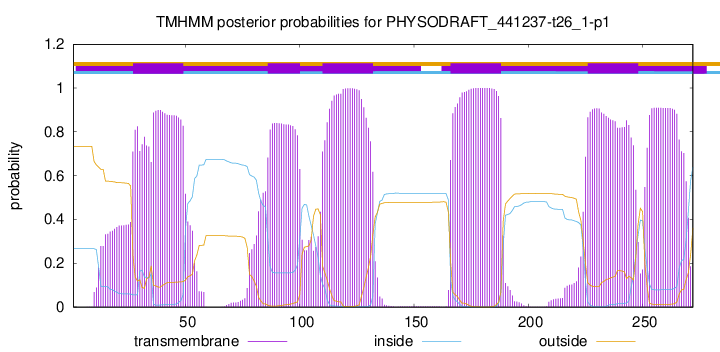
| Start | End |
|---|---|
| 27 | 49 |
| 86 | 100 |
| 110 | 132 |
| 166 | 188 |
| 226 | 248 |
