You are browsing environment: FUNGIDB
PHYSODRAFT_362341-t26_1-p1
Basic Information
help
Species
Phytophthora sojae
Lineage
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae
CAZyme ID
PHYSODRAFT_362341-t26_1-p1
CAZy Family
GH3
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
411
44538.68
4.1033
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_PsojaeP6497
28142
1094619
1653
26489
Gene Location
No EC number prediction in PHYSODRAFT_362341-t26_1-p1.
Family
Start
End
Evalue
family coverage
GH72
64
255
3.5e-32
0.5160256410256411
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
397351
Glyco_hydro_72
1.98e-16
66
267
127
303
Glucanosyltransferase. This is a family of glycosylphosphatidylinositol-anchored beta(1-3)glucanosyltransferases. The active site residues in the Aspergillus fumigatus example are the two glutamate residues at 160 and 261.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
4.47e-130
18
397
27
477
6.74e-104
63
358
149
440
1.62e-103
137
360
2
218
3.23e-103
19
358
42
456
1.40e-84
54
360
2302
2595
PHYSODRAFT_362341-t26_1-p1 has no PDB hit.
PHYSODRAFT_362341-t26_1-p1 has no Swissprot hit.
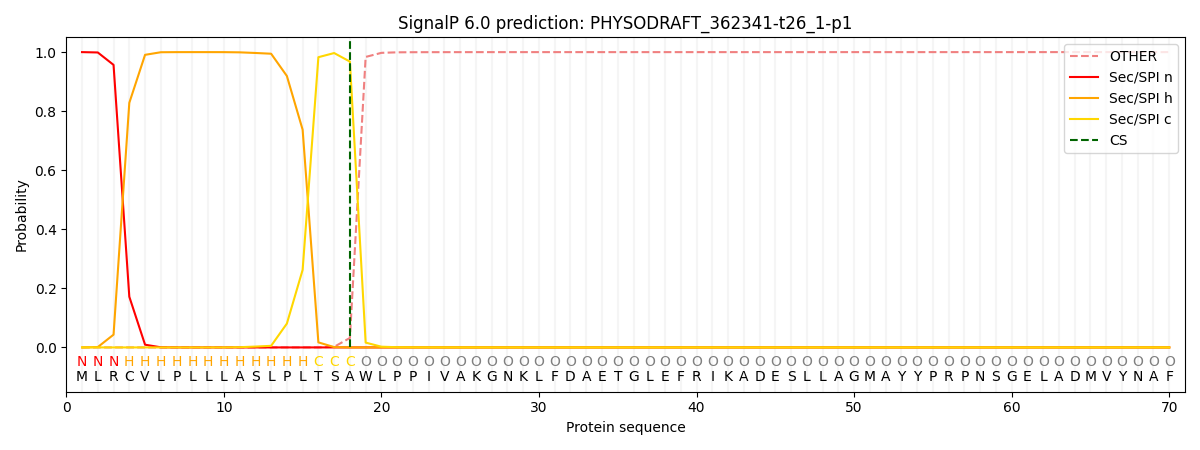
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000332
0.999613
CS pos: 18-19. Pr: 0.9679
There is no transmembrane helices in PHYSODRAFT_362341-t26_1-p1.