You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_354534-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_354534-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
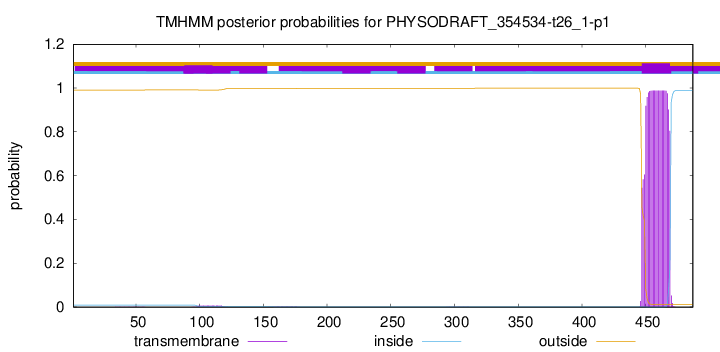
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_354534-t26_1-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:2 | 3.2.1.23:1 | 3.2.1.38:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 1 | 424 | 5.5e-101 | 0.8344988344988346 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 2.26e-102 | 1 | 425 | 67 | 450 | Glycosyl hydrolase family 1. |
| 225343 | BglB | 2.27e-85 | 1 | 430 | 68 | 457 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215455 | PLN02849 | 5.34e-67 | 1 | 419 | 88 | 476 | beta-glucosidase |
| 215435 | PLN02814 | 1.31e-61 | 1 | 419 | 86 | 476 | beta-glucosidase |
| 184102 | PRK13511 | 4.42e-52 | 3 | 426 | 65 | 466 | 6-phospho-beta-galactosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.71e-208 | 1 | 484 | 72 | 562 | |
| 4.88e-204 | 1 | 468 | 91 | 564 | |
| 4.41e-158 | 1 | 434 | 82 | 512 | |
| 7.60e-142 | 1 | 429 | 16 | 419 | |
| 1.27e-72 | 1 | 421 | 108 | 513 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.71e-68 | 5 | 394 | 79 | 445 | Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8],4GXP_B Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8],4GXP_C Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8] |
|
| 5.69e-67 | 1 | 422 | 82 | 478 | Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],2RGL_B Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],2RGM_A Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],2RGM_B Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],7BZM_A Crystal structure of rice Os3BGlu7 with glucoimidazole [Oryza sativa Japonica Group],7BZM_B Crystal structure of rice Os3BGlu7 with glucoimidazole [Oryza sativa Japonica Group] |
|
| 1.56e-66 | 1 | 422 | 82 | 478 | Crystal structure of rice BGlu1 E176Q/Y341A mutant complexed with cellotetraose [Oryza sativa Japonica Group],4QLK_B Crystal structure of rice BGlu1 E176Q/Y341A mutant complexed with cellotetraose [Oryza sativa Japonica Group] |
|
| 1.56e-66 | 1 | 422 | 82 | 478 | Crystal structure of rice BGlu1 E176Q mutant in complex with laminaribiose [Oryza sativa Japonica Group],3AHT_B Crystal structure of rice BGlu1 E176Q mutant in complex with laminaribiose [Oryza sativa Japonica Group],3AHV_A Semi-active E176Q mutant of rice bglu1 covalent complex with 2-deoxy-2-fluoroglucoside [Oryza sativa Japonica Group],3AHV_B Semi-active E176Q mutant of rice bglu1 covalent complex with 2-deoxy-2-fluoroglucoside [Oryza sativa Japonica Group],3F4V_A Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],3F4V_B Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],3F5J_A Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],3F5J_B Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],3F5K_A Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],3F5K_B Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],3F5L_A Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group],3F5L_B Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase [Oryza sativa Japonica Group] |
|
| 2.19e-66 | 1 | 422 | 82 | 478 | Crystal Structure of Rice BGlu1 E386G/S334A Mutant Complexed with Cellotetraose [Oryza sativa Japonica Group],3SCV_B Crystal Structure of Rice BGlu1 E386G/S334A Mutant Complexed with Cellotetraose [Oryza sativa Japonica Group] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.81e-68 | 6 | 421 | 115 | 507 | Myrosinase 5 OS=Arabidopsis thaliana OX=3702 GN=TGG5 PE=1 SV=1 |
|
| 7.44e-68 | 1 | 421 | 81 | 487 | Beta-glucosidase 27 OS=Arabidopsis thaliana OX=3702 GN=BGLU27 PE=2 SV=2 |
|
| 3.17e-67 | 1 | 434 | 82 | 503 | Beta-glucosidase 26, peroxisomal OS=Arabidopsis thaliana OX=3702 GN=BGLU26 PE=1 SV=1 |
|
| 1.53e-66 | 1 | 416 | 73 | 483 | Beta-glucosidase 1B OS=Phanerodontia chrysosporium OX=2822231 GN=BGL1B PE=1 SV=1 |
|
| 1.55e-66 | 6 | 421 | 115 | 507 | Myrosinase 4 OS=Arabidopsis thaliana OX=3702 GN=TGG4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000030 | 0.000000 |