You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_335255-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_335255-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_335255-t26_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | flavodoxin/amb allergen/pectate lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.10:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 178 | 282 | 2e-40 | 0.5454545454545454 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 1.08e-18 | 122 | 283 | 2 | 189 | Amb_all domain. |
| 226384 | PelB | 3.12e-08 | 97 | 323 | 51 | 314 | Pectate lyase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.96e-232 | 1 | 361 | 1 | 396 | |
| 2.08e-227 | 1 | 361 | 1 | 396 | |
| 8.92e-180 | 1 | 361 | 1 | 396 | |
| 6.45e-163 | 42 | 361 | 133 | 487 | |
| 2.50e-143 | 10 | 361 | 10 | 406 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.11e-69 | 24 | 360 | 4 | 358 | Pectin Lyase B [Aspergillus niger] |
|
| 1.95e-65 | 25 | 323 | 5 | 321 | Pectin Lyase A [Aspergillus niger] |
|
| 3.33e-63 | 25 | 323 | 5 | 321 | Pectin Lyase A [Aspergillus niger],1IDJ_B Pectin Lyase A [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.10e-75 | 1 | 323 | 1 | 341 | Pectin lyase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pelB PE=2 SV=1 |
|
| 2.76e-75 | 13 | 349 | 12 | 369 | Probable pectin lyase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pelB PE=3 SV=2 |
|
| 4.25e-75 | 10 | 323 | 8 | 340 | Probable pectin lyase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=pelA PE=3 SV=1 |
|
| 4.88e-74 | 25 | 323 | 24 | 341 | Probable pectin lyase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pelA PE=3 SV=1 |
|
| 2.65e-73 | 1 | 325 | 1 | 342 | Probable pectin lyase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pelA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
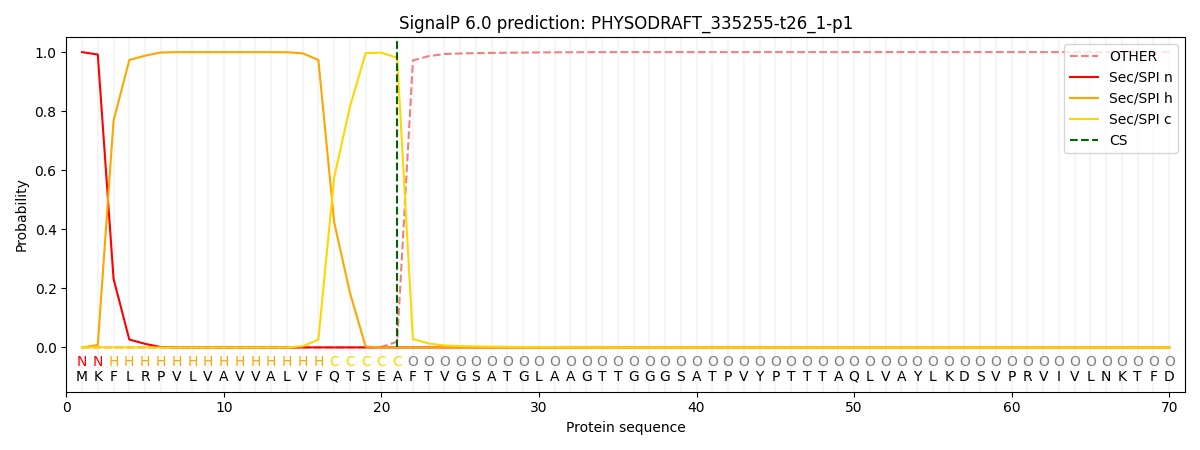
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000198 | 0.999747 | CS pos: 21-22. Pr: 0.9806 |
