You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_326784-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_326784-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_326784-t26_1-p1 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT62 | 192 | 405 | 1.1e-31 | 0.8619402985074627 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397491 | Anp1 | 2.93e-29 | 221 | 405 | 38 | 245 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
| 411474 | fibronec_FbpA | 3.01e-05 | 30 | 136 | 156 | 261 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
| 411384 | Streccoc_I_II | 3.91e-05 | 63 | 152 | 893 | 984 | antigen I/II family LPXTG-anchored adhesin. Members of the antigen I/II family are adhesins with a glucan-binding domain, two types of repetitive regions, an isopeptide bond-forming domain associated with shear resistance, and a C-terminal LPXTG motif for anchoring to the cell wall. They occur in oral Streptococci, and tend to be major cell surface adhesins. Members of this family include SspA and SspB from Streptococcus gordonii, antigen I/II from S. mutans, etc. |
| 380203 | Vip_LPXTG_Lm | 4.75e-05 | 28 | 147 | 243 | 346 | cell invasion LPXTG protein Vip. Vip (Virulence protein), like the LPXTG-type internalins, is an LPXTG-anchored surface protein of the mammalian cell-invading pathogen Listeria monocytogenes, but absent from the related species Listeria innocua. For certain cell types, Vip is required for Listeria's ability to invade. It appears to bind the endoplasmic reticulum (ER) resident chaperone Gp96 as its receptor. |
| 139494 | PRK13335 | 8.28e-05 | 10 | 170 | 11 | 186 | superantigen-like protein SSL3; Reviewed. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.12e-30 | 191 | 435 | 77 | 368 | |
| 9.12e-30 | 191 | 435 | 77 | 368 | |
| 2.16e-27 | 193 | 435 | 95 | 384 | |
| 2.16e-27 | 193 | 435 | 95 | 384 | |
| 2.16e-27 | 193 | 435 | 95 | 384 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.05e-08 | 222 | 435 | 40 | 292 | Crystal structure of Saccharomyces cerevisiae Mnn9 in complex with GDP and Mn. [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.22e-15 | 230 | 437 | 89 | 329 | Mannan polymerase complex subunit mnn9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mnn9 PE=3 SV=1 |
|
| 9.82e-15 | 222 | 435 | 116 | 364 | Mannan polymerase II complex anp1 subunit OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=anp1 PE=3 SV=1 |
|
| 2.78e-11 | 222 | 380 | 99 | 271 | Mannan polymerase II complex ANP1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ANP1 PE=1 SV=3 |
|
| 1.17e-09 | 267 | 405 | 309 | 460 | Mannan polymerase I complex VAN1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=VAN1 PE=1 SV=3 |
|
| 3.19e-09 | 181 | 435 | 75 | 358 | Mannan polymerase complex subunit MNN9 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN9 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
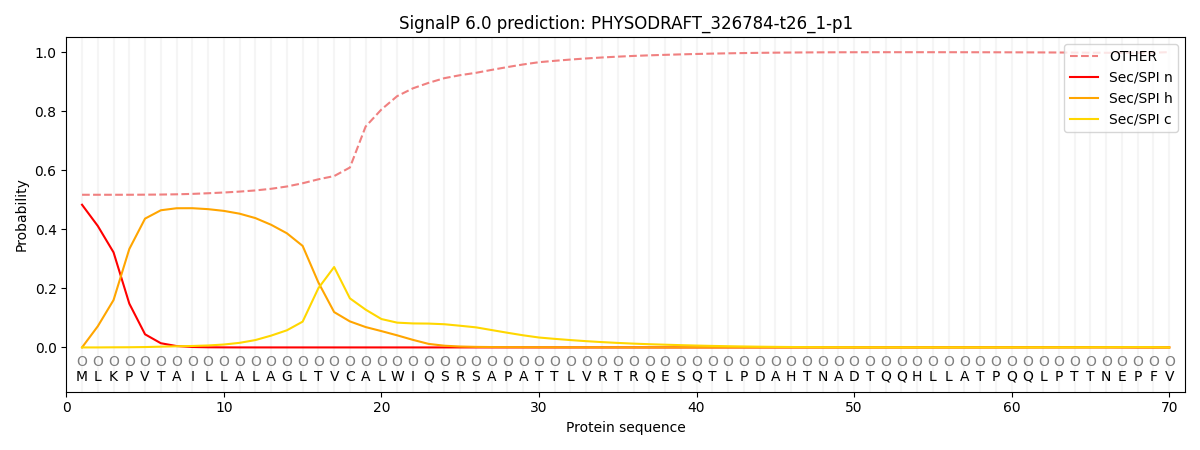
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.521743 | 0.478262 |
