You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_324715-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_324715-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_324715-t26_1-p1 | |||||||||||
| CAZy Family | GH1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.-:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 25 | 257 | 6.7e-46 | 0.9510204081632653 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 411470 | opacity_OapA | 3.07e-04 | 191 | 265 | 189 | 262 | opacity-associated protein OapA. This family consists of full-length homologs to OapA, opacity-associated protein A as described in Haemophilus influenzae. OapA shares a C-terminal homology domain, called the OapA domain, with the Escherichia coli protein YtfB, which is now known to bind peptidoglycan through its OapA domain and to act as a cell division protein. |
| 237019 | PRK11907 | 0.001 | 186 | 262 | 8 | 82 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| 236776 | PRK10856 | 0.003 | 195 | 263 | 151 | 220 | cytoskeleton protein RodZ. |
| 237030 | kgd | 0.009 | 182 | 264 | 30 | 107 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.04e-76 | 13 | 266 | 3 | 246 | |
| 2.23e-74 | 13 | 266 | 3 | 250 | |
| 5.48e-72 | 13 | 220 | 3 | 205 | |
| 2.11e-55 | 19 | 200 | 6 | 203 | |
| 2.81e-55 | 19 | 206 | 6 | 209 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.30e-44 | 71 | 193 | 55 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
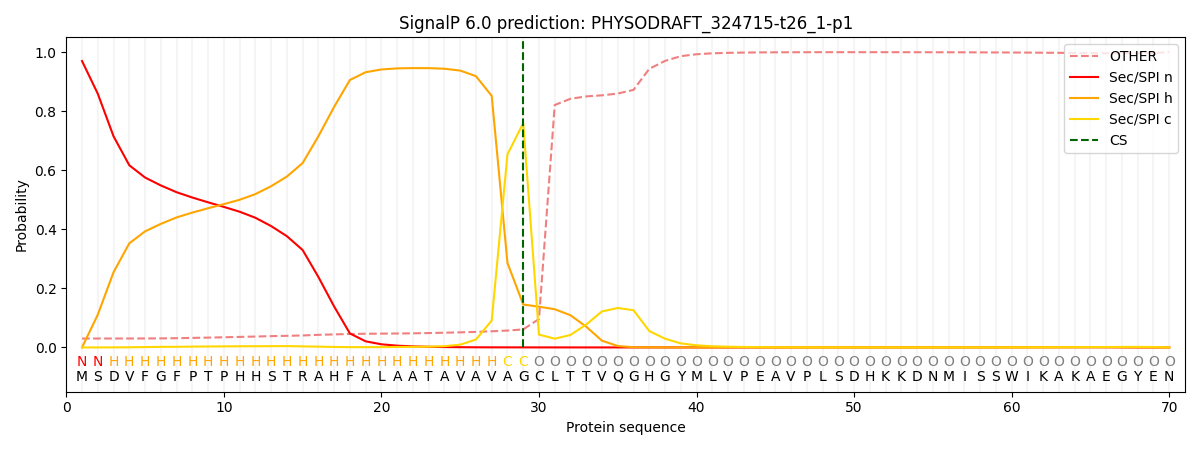
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.032361 | 0.967624 | CS pos: 29-30. Pr: 0.7594 |
