You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_263403-t26_1-p1
Basic Information
help
| Species |
Phytophthora sojae
|
| Lineage |
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae
|
| CAZyme ID |
PHYSODRAFT_263403-t26_1-p1
|
| CAZy Family |
PL3|PL3 |
| CAZyme Description |
putative endo-1,3-beta glucanase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 359 |
|
40436.63 |
4.6752 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PsojaeP6497 |
28142 |
1094619 |
1653 |
26489
|
|
| Gene Location |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 227625
|
Scw11 |
1.49e-24 |
2 |
274 |
5 |
302 |
Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 395527
|
Ricin_B_lectin |
0.001 |
324 |
351 |
77 |
104 |
Ricin-type beta-trefoil lectin domain. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 5.51e-119 |
1 |
351 |
1 |
371 |
| 5.51e-119 |
1 |
351 |
1 |
371 |
| 5.51e-119 |
1 |
351 |
1 |
371 |
| 5.51e-119 |
1 |
351 |
1 |
371 |
| 5.51e-119 |
1 |
351 |
1 |
371 |
PHYSODRAFT_263403-t26_1-p1 has no PDB hit.
PHYSODRAFT_263403-t26_1-p1 has no Swissprot hit.
This protein is predicted as SP
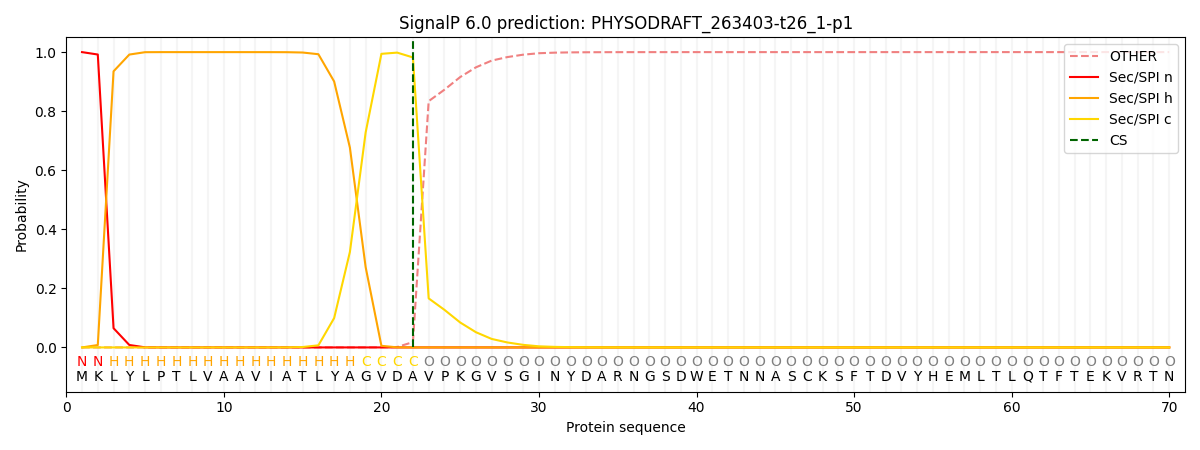
| Other |
SP_Sec_SPI |
CS Position |
| 0.000191 |
0.999761 |
CS pos: 22-23. Pr: 0.9819 |
There is no transmembrane helices in PHYSODRAFT_263403-t26_1-p1.

