You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_246985-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_246985-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_246985-t26_1-p1 | |||||||||||
| CAZy Family | AA16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:2 | 3.2.1.23:1 | 3.2.1.38:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 41 | 522 | 2e-131 | 0.9813519813519813 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 9.55e-139 | 42 | 523 | 5 | 450 | Glycosyl hydrolase family 1. |
| 274539 | BGL | 1.81e-114 | 43 | 517 | 1 | 426 | beta-galactosidase. |
| 225343 | BglB | 1.99e-113 | 42 | 531 | 4 | 460 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215435 | PLN02814 | 5.61e-81 | 42 | 491 | 28 | 462 | beta-glucosidase |
| 215455 | PLN02849 | 4.22e-76 | 42 | 518 | 30 | 477 | beta-glucosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.27e-203 | 41 | 531 | 31 | 520 | |
| 6.51e-202 | 41 | 527 | 22 | 507 | |
| 2.39e-199 | 41 | 530 | 12 | 500 | |
| 2.13e-121 | 91 | 527 | 5 | 419 | |
| 1.36e-97 | 42 | 522 | 4 | 454 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.44e-92 | 42 | 519 | 43 | 504 | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine [Rauvolfia serpentina],2JF6_B Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine [Rauvolfia serpentina],2JF7_A Structure of Strictosidine Glucosidase [Rauvolfia serpentina],2JF7_B Structure of Strictosidine Glucosidase [Rauvolfia serpentina],3ZJ7_A Crystal structure of strictosidine glucosidase in complex with inhibitor-1 [Rauvolfia serpentina],3ZJ7_B Crystal structure of strictosidine glucosidase in complex with inhibitor-1 [Rauvolfia serpentina],3ZJ8_A Crystal structure of strictosidine glucosidase in complex with inhibitor-2 [Rauvolfia serpentina],3ZJ8_B Crystal structure of strictosidine glucosidase in complex with inhibitor-2 [Rauvolfia serpentina] |
|
| 1.79e-88 | 40 | 529 | 9 | 453 | Chain A, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1H_B Chain B, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1M_A Chain A, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_B Chain B, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_C Chain C, Ancestral reconstructed glycosidase [synthetic construct] |
|
| 1.22e-86 | 42 | 502 | 22 | 505 | Crystal structure of native Raucaffricine glucosidase [Rauvolfia serpentina],4ATD_B Crystal structure of native Raucaffricine glucosidase [Rauvolfia serpentina],4ATL_A Crystal structure of Raucaffricine glucosidase in complex with Glucose [Rauvolfia serpentina],4ATL_B Crystal structure of Raucaffricine glucosidase in complex with Glucose [Rauvolfia serpentina] |
|
| 2.05e-86 | 42 | 502 | 39 | 499 | Crystal structure of beta-primeverosidase [Camellia sinensis],3WQ4_B Crystal structure of beta-primeverosidase [Camellia sinensis],3WQ5_A beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, natural aglycone derivative [Camellia sinensis],3WQ5_B beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, natural aglycone derivative [Camellia sinensis],3WQ6_A beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, artificial aglycone derivative [Camellia sinensis],3WQ6_B beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, artificial aglycone derivative [Camellia sinensis] |
|
| 2.49e-86 | 42 | 502 | 22 | 505 | Crystal of Raucaffricine Glucosidase in complex with inhibitor [Rauvolfia serpentina],3ZJ6_B Crystal of Raucaffricine Glucosidase in complex with inhibitor [Rauvolfia serpentina],4A3Y_A Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway [Rauvolfia serpentina],4A3Y_B Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway [Rauvolfia serpentina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.71e-92 | 42 | 491 | 31 | 485 | Beta-glucosidase 30 OS=Arabidopsis thaliana OX=3702 GN=BGLU30 PE=2 SV=1 |
|
| 6.79e-92 | 42 | 502 | 19 | 486 | Beta-glucosidase 26, peroxisomal OS=Arabidopsis thaliana OX=3702 GN=BGLU26 PE=1 SV=1 |
|
| 1.25e-91 | 42 | 519 | 43 | 504 | Strictosidine-O-beta-D-glucosidase OS=Rauvolfia serpentina OX=4060 GN=SGR1 PE=1 SV=1 |
|
| 2.62e-91 | 42 | 502 | 37 | 503 | Beta-glucosidase 31 OS=Arabidopsis thaliana OX=3702 GN=BGLU31 PE=2 SV=1 |
|
| 2.23e-90 | 42 | 488 | 32 | 478 | Beta-glucosidase 30 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU30 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
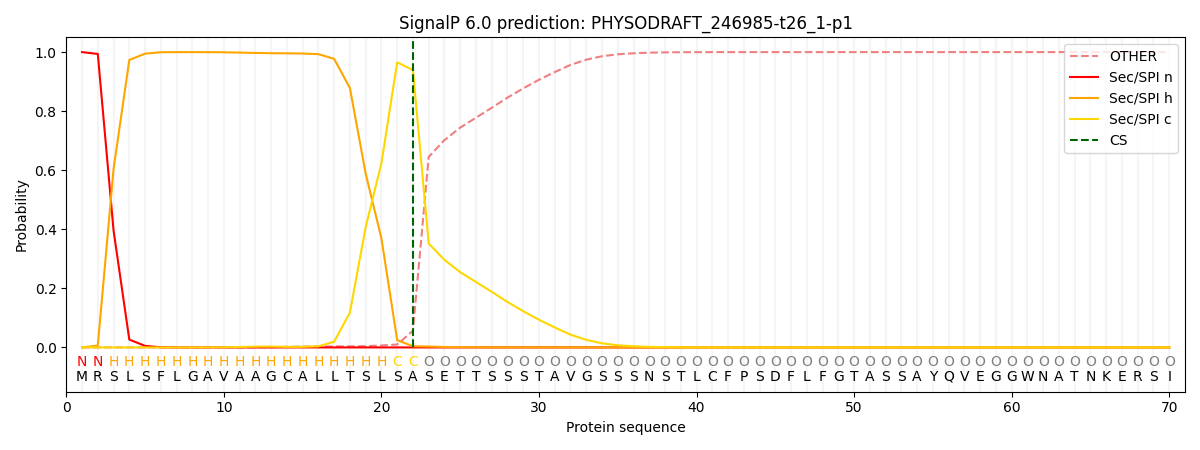
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000659 | 0.999306 | CS pos: 22-23. Pr: 0.9395 |
