You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_97674T0-p1
You are here: Home > Sequence: PHYCI_97674T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_97674T0-p1 | |||||||||||
| CAZy Family | GT71 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 21 | 274 | 1.7e-17 | 0.9678456591639871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 5.13e-14 | 21 | 233 | 47 | 262 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 311610 | Glyco_hydro_53 | 3.84e-04 | 104 | 248 | 126 | 295 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.57e-102 | 3 | 281 | 13 | 288 | |
| 2.35e-89 | 21 | 283 | 574 | 836 | |
| 1.27e-68 | 21 | 281 | 29 | 288 | |
| 3.71e-51 | 21 | 194 | 29 | 202 | |
| 5.73e-41 | 21 | 281 | 22 | 282 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.86e-12 | 34 | 216 | 326 | 508 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
|
| 1.86e-12 | 34 | 216 | 326 | 508 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 5.72e-12 | 21 | 216 | 297 | 495 | Probable beta-glucosidase btgE OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
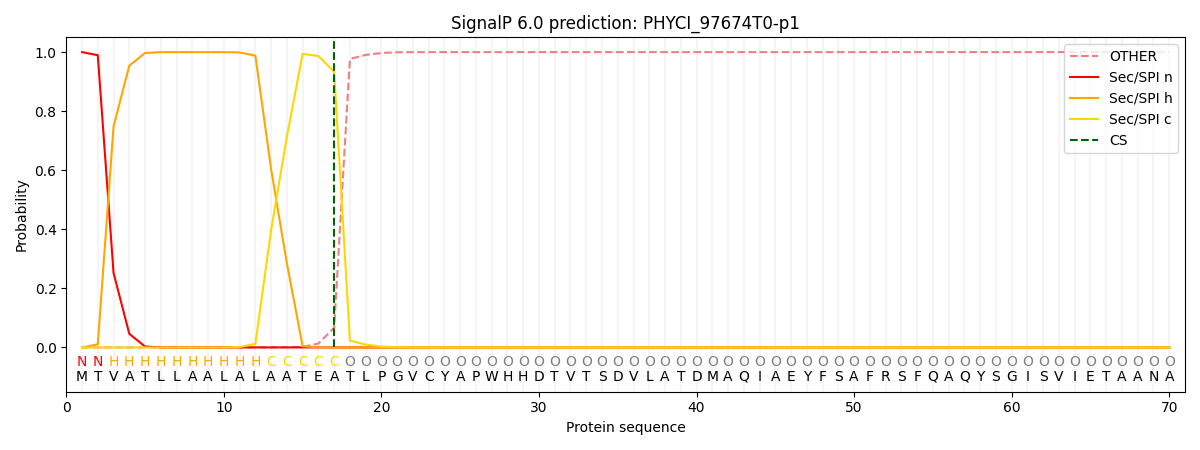
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000247 | 0.999735 | CS pos: 17-18. Pr: 0.9332 |
