You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_94152T0-p1
You are here: Home > Sequence: PHYCI_94152T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_94152T0-p1 | |||||||||||
| CAZy Family | GT62 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 79 | 270 | 7.8e-39 | 0.9896907216494846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397360 | Pectate_lyase | 1.85e-36 | 75 | 270 | 4 | 195 | Pectate lyase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.48e-107 | 11 | 298 | 10 | 283 | |
| 6.62e-43 | 73 | 295 | 24 | 252 | |
| 1.66e-41 | 60 | 294 | 21 | 259 | |
| 5.47e-39 | 73 | 293 | 34 | 258 | |
| 4.46e-33 | 75 | 252 | 22 | 202 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.52e-09 | 124 | 270 | 36 | 196 | Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B90_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi] |
|
| 7.62e-09 | 124 | 270 | 154 | 314 | Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B4N_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi],3B8Y_A Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B8Y_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.91e-33 | 76 | 270 | 28 | 219 | Probable pectate lyase G OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyG PE=3 SV=1 |
|
| 3.48e-31 | 76 | 270 | 43 | 236 | Probable pectate lyase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyD PE=3 SV=1 |
|
| 1.03e-30 | 76 | 289 | 35 | 247 | Probable pectate lyase D OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyD PE=3 SV=1 |
|
| 1.06e-30 | 50 | 276 | 13 | 236 | Probable pectate lyase E OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=plyE PE=3 SV=1 |
|
| 1.84e-28 | 76 | 270 | 41 | 232 | Pectate lyase H OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
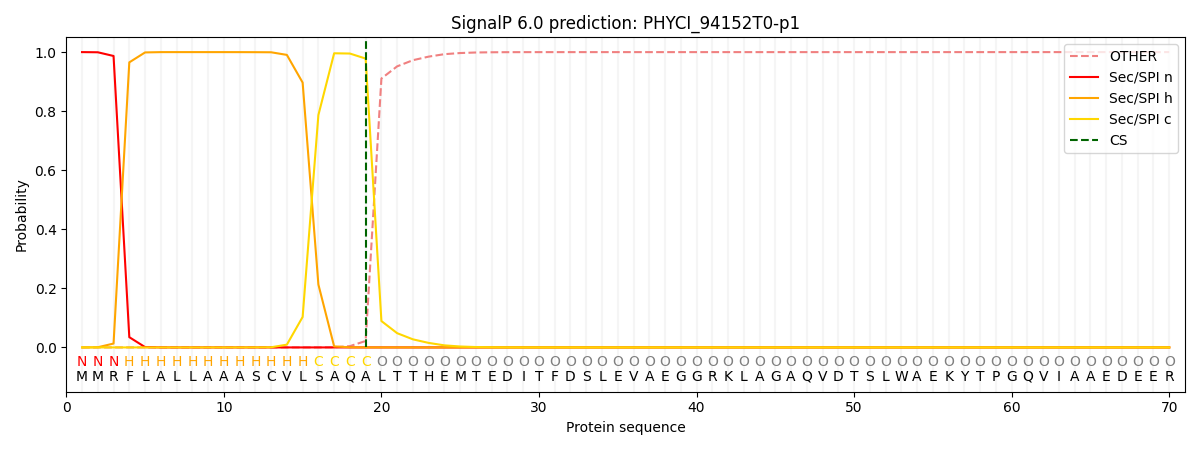
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000225 | 0.999734 | CS pos: 19-20. Pr: 0.9781 |
