You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_82515T0-p1
You are here: Home > Sequence: PHYCI_82515T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_82515T0-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.-:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 11 | 276 | 6.7e-64 | 0.9510204081632653 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 411525 | agg_sub_LPXTH | 2.72e-06 | 198 | 309 | 72 | 185 | serine-rich aggregation substance UasX. Members of this protein family are repetitive, serine-rich surface proteins of the Firmicutes, found primarily in the genus Leuconostoc. The variant form of sortase signal, LPXTH, is replaced by LPXTG in members from some lineages, such as Weissella oryzae, and therefore recognizable. Some members of this family have the KxYKxGKxW type signal peptide as seen in the glycoprotein adhesin GspB, a substrate of the accessory Sec system for secretion. WOSG25_050600 from Weissella oryzae SG25 is identified in a publication as an unnamed aggregation substance, a conclusion supported by the sorting signals and composition reported here. We assign the gene symbol uasX (unnamed aggregation substance X) based on our evaluation of the family. |
| 380146 | MSCRAMM_SdrC | 2.32e-05 | 201 | 315 | 824 | 939 | MSCRAMM family adhesin SdrC. Features of this protein family include a YSIRK-type signal peptide at the N-terminus and a variable-length C-terminal region of Ser-Asp (SD) repeats followed by an LPXTG motif for surface immobilization by sortase. |
| 215130 | PLN02217 | 5.56e-05 | 197 | 290 | 563 | 652 | probable pectinesterase/pectinesterase inhibitor |
| 223039 | PHA03307 | 7.06e-05 | 195 | 321 | 276 | 402 | transcriptional regulator ICP4; Provisional |
| 215130 | PLN02217 | 1.12e-04 | 208 | 304 | 563 | 651 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.32e-153 | 1 | 319 | 1 | 319 | |
| 8.60e-140 | 1 | 213 | 1 | 213 | |
| 3.49e-139 | 1 | 213 | 1 | 213 | |
| 9.16e-135 | 1 | 213 | 1 | 213 | |
| 1.48e-121 | 1 | 319 | 1 | 275 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.84e-116 | 24 | 195 | 1 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
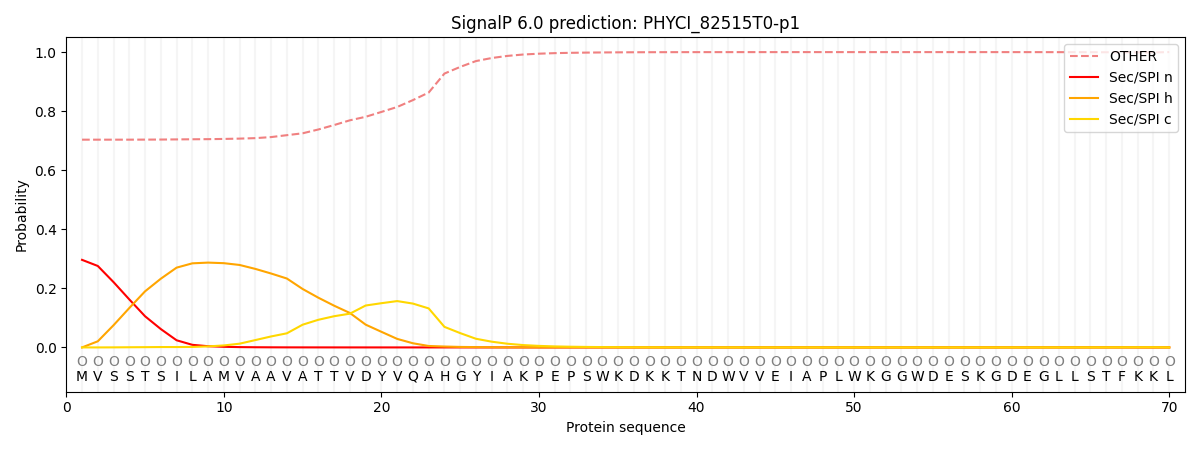
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.716730 | 0.283268 |
