You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_66922T0-p1
You are here: Home > Sequence: PHYCI_66922T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
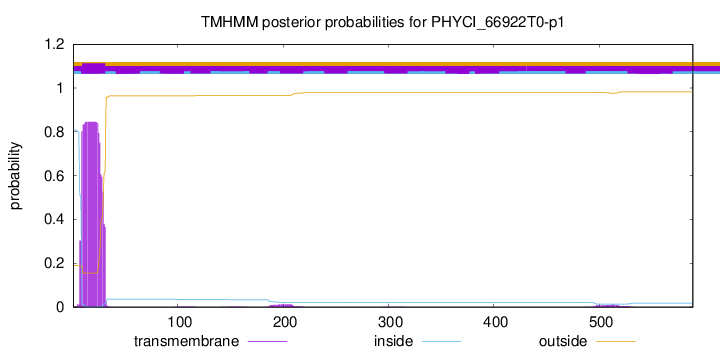
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_66922T0-p1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | Stationary phase-induced protein, SOR/SNZ family | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 179769 | PRK04180 | 0.0 | 296 | 573 | 5 | 284 | pyridoxal 5'-phosphate synthase lyase subunit PdxS. |
| 223292 | PdxS | 0.0 | 296 | 568 | 8 | 280 | Pyridoxal biosynthesis lyase PdxS [Coenzyme transport and metabolism]. |
| 240078 | pdxS | 0.0 | 302 | 568 | 2 | 268 | PdxS is a subunit of the pyridoxal 5'-phosphate (PLP) synthase, an important enzyme in deoxyxylulose 5-phosphate (DXP)-independent pathway for de novo biosynthesis of PLP, present in some eubacteria, in archaea, fungi, plants, plasmodia, and some metazoa. Together with PdxT, PdxS forms the PLP synthase, a heteromeric glutamine amidotransferase (GATase), whereby PdxT produces ammonia from glutamine and PdxS combines ammonia with five- and three-carbon phosphosugars to form PLP. PLP is the biologically active form of vitamin B6, an essential cofactor in many biochemical processes. PdxS subunits form two hexameric rings. |
| 129443 | TIGR00343 | 8.21e-170 | 299 | 568 | 1 | 271 | pyridoxal 5'-phosphate synthase, synthase subunit Pdx1. This protein had been believed to be a singlet oxygen resistance protein. Subsequent work showed that it is a protein of pyridoxine (vitamin B6) biosynthesis, and that pyridoxine quenches the highly toxic singlet form of oxygen produced by light in the presence of certain chemicals. [Biosynthesis of cofactors, prosthetic groups, and carriers, Pyridoxine] |
| 396308 | SOR_SNZ | 1.23e-159 | 296 | 501 | 1 | 206 | SOR/SNZ family. Members of this family are enzymes involved in a new pathway of pyridoxine/pyridoxal 5-phosphate biosynthesis. This family was formerly known as UPF0019. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.94e-33 | 76 | 283 | 11 | 226 | |
| 5.94e-33 | 76 | 283 | 11 | 226 | |
| 4.68e-30 | 79 | 291 | 7 | 216 | |
| 6.58e-30 | 79 | 269 | 2 | 188 | |
| 2.60e-29 | 74 | 279 | 2 | 204 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.13e-143 | 293 | 568 | 19 | 294 | PDX1.3-adduct (Arabidopsis) [Arabidopsis thaliana],5K2Z_B PDX1.3-adduct (Arabidopsis) [Arabidopsis thaliana],5K2Z_C PDX1.3-adduct (Arabidopsis) [Arabidopsis thaliana],5K2Z_D PDX1.3-adduct (Arabidopsis) [Arabidopsis thaliana],5K3V_A apo-PDX1.3 (Arabidopsis) [Arabidopsis thaliana],5K3V_B apo-PDX1.3 (Arabidopsis) [Arabidopsis thaliana],5K3V_C apo-PDX1.3 (Arabidopsis) [Arabidopsis thaliana],5K3V_D apo-PDX1.3 (Arabidopsis) [Arabidopsis thaliana] |
|
| 3.24e-143 | 293 | 568 | 19 | 294 | Crystal structure of Arabidopsis thaliana Pdx1-PLP complex [Arabidopsis thaliana],5LNR_B Crystal structure of Arabidopsis thaliana Pdx1-PLP complex [Arabidopsis thaliana],5LNR_C Crystal structure of Arabidopsis thaliana Pdx1-PLP complex [Arabidopsis thaliana],5LNR_D Crystal structure of Arabidopsis thaliana Pdx1-PLP complex [Arabidopsis thaliana],5LNS_A Crystal structure of Arabidopsis thaliana Pdx1-R5P complex [Arabidopsis thaliana],5LNS_B Crystal structure of Arabidopsis thaliana Pdx1-R5P complex [Arabidopsis thaliana],5LNS_C Crystal structure of Arabidopsis thaliana Pdx1-R5P complex [Arabidopsis thaliana],5LNS_D Crystal structure of Arabidopsis thaliana Pdx1-R5P complex [Arabidopsis thaliana],5LNU_A Crystal structure of Arabidopsis thaliana Pdx1-I320 complex [Arabidopsis thaliana],5LNU_B Crystal structure of Arabidopsis thaliana Pdx1-I320 complex [Arabidopsis thaliana],5LNU_C Crystal structure of Arabidopsis thaliana Pdx1-I320 complex [Arabidopsis thaliana],5LNU_D Crystal structure of Arabidopsis thaliana Pdx1-I320 complex [Arabidopsis thaliana],5LNV_A Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals [Arabidopsis thaliana],5LNV_B Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals [Arabidopsis thaliana],5LNV_C Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals [Arabidopsis thaliana],5LNV_D Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals [Arabidopsis thaliana],5LNW_A Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex [Arabidopsis thaliana],5LNW_B Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex [Arabidopsis thaliana],5LNW_C Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex [Arabidopsis thaliana],5LNW_D Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex [Arabidopsis thaliana] |
|
| 7.03e-143 | 293 | 568 | 48 | 323 | Chain M, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_N Chain N, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_O Chain O, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_P Chain P, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_Q Chain Q, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_R Chain R, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_S Chain S, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_T Chain T, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_U Chain U, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_V Chain V, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_W Chain W, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana],7LB6_X Chain X, Pyridoxal 5'-phosphate synthase subunit PDX1.3 [Arabidopsis thaliana] |
|
| 2.52e-142 | 293 | 568 | 19 | 294 | PDX1.2/PDX1.3 complex (PDX1.3:K97A) [Arabidopsis thaliana],6HYE_D PDX1.2/PDX1.3 complex (PDX1.3:K97A) [Arabidopsis thaliana],6HYE_F PDX1.2/PDX1.3 complex (PDX1.3:K97A) [Arabidopsis thaliana],6HYE_H PDX1.2/PDX1.3 complex (PDX1.3:K97A) [Arabidopsis thaliana] |
|
| 5.24e-142 | 292 | 568 | 19 | 295 | Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex [Arabidopsis thaliana],5LNT_B Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex [Arabidopsis thaliana],5LNT_C Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex [Arabidopsis thaliana],5LNT_D Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.23e-150 | 296 | 569 | 5 | 278 | Pyridoxal 5'-phosphate synthase subunit PdxS OS=Roseiflexus sp. (strain RS-1) OX=357808 GN=pdxS PE=3 SV=1 |
|
| 1.17e-149 | 296 | 568 | 5 | 277 | Pyridoxal 5'-phosphate synthase subunit PdxS OS=Chloroflexus aurantiacus (strain ATCC 29364 / DSM 637 / Y-400-fl) OX=480224 GN=pdxS PE=3 SV=1 |
|
| 1.17e-149 | 296 | 568 | 5 | 277 | Pyridoxal 5'-phosphate synthase subunit PdxS OS=Chloroflexus aurantiacus (strain ATCC 29366 / DSM 635 / J-10-fl) OX=324602 GN=pdxS PE=3 SV=1 |
|
| 1.33e-148 | 296 | 568 | 5 | 277 | Pyridoxal 5'-phosphate synthase subunit PdxS OS=Chloroflexus aggregans (strain MD-66 / DSM 9485) OX=326427 GN=pdxS PE=3 SV=1 |
|
| 7.60e-148 | 296 | 569 | 5 | 278 | Pyridoxal 5'-phosphate synthase subunit PdxS OS=Roseiflexus castenholzii (strain DSM 13941 / HLO8) OX=383372 GN=pdxS PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
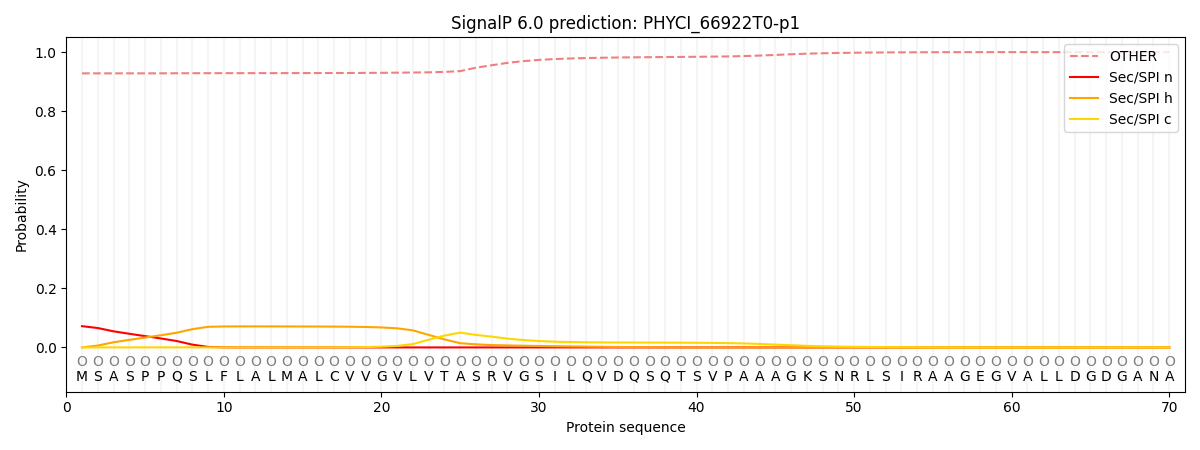
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.931655 | 0.068383 |