You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_251762T0-p1
You are here: Home > Sequence: PHYCI_251762T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
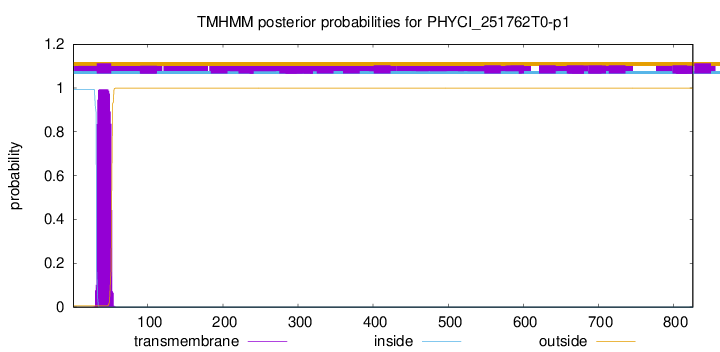
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_251762T0-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 92 | 393 | 9.9e-88 | 0.993485342019544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 166698 | PLN03059 | 6.24e-121 | 84 | 797 | 29 | 728 | beta-galactosidase; Provisional |
| 396048 | Glyco_hydro_35 | 5.38e-95 | 92 | 392 | 2 | 315 | Glycosyl hydrolases family 35. |
| 224786 | GanA | 8.40e-21 | 85 | 357 | 1 | 292 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 396834 | Glyco_hydro_42 | 1.74e-05 | 116 | 251 | 12 | 150 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 4 | 798 | 3 | 796 | |
| 2.32e-245 | 28 | 797 | 39 | 794 | |
| 2.81e-151 | 83 | 786 | 32 | 721 | |
| 1.04e-147 | 79 | 793 | 28 | 732 | |
| 8.11e-147 | 83 | 786 | 32 | 721 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.30e-103 | 83 | 796 | 6 | 706 | Chain A, Beta-galactosidase [Solanum lycopersicum],3W5F_B Chain B, Beta-galactosidase [Solanum lycopersicum],3W5G_A Chain A, Beta-galactosidase [Solanum lycopersicum],3W5G_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK5_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK5_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
|
| 5.42e-102 | 83 | 796 | 6 | 706 | Chain A, Beta-galactosidase [Solanum lycopersicum],6IK6_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK7_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK7_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK8_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK8_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
|
| 7.71e-44 | 92 | 357 | 24 | 294 | Chain A, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
|
| 1.38e-36 | 82 | 362 | 17 | 311 | Galactanase BT0290 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 3.01e-36 | 83 | 368 | 16 | 308 | Chain A, Beta-galactosidase [Mus musculus],7KDV_C Chain C, Beta-galactosidase [Mus musculus],7KDV_E Chain E, Beta-galactosidase [Mus musculus],7KDV_G Chain G, Beta-galactosidase [Mus musculus],7KDV_I Chain I, Beta-galactosidase [Mus musculus],7KDV_K Chain K, Beta-galactosidase [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.73e-110 | 84 | 797 | 30 | 731 | Beta-galactosidase 5 OS=Arabidopsis thaliana OX=3702 GN=BGAL5 PE=2 SV=1 |
|
| 3.92e-107 | 84 | 792 | 26 | 721 | Beta-galactosidase 5 OS=Oryza sativa subsp. japonica OX=39947 GN=Os03g0165400 PE=2 SV=1 |
|
| 4.04e-106 | 85 | 797 | 33 | 733 | Beta-galactosidase 3 OS=Arabidopsis thaliana OX=3702 GN=BGAL3 PE=2 SV=1 |
|
| 1.14e-105 | 69 | 792 | 22 | 724 | Beta-galactosidase 4 OS=Oryza sativa subsp. indica OX=39946 GN=OsI_006270 PE=3 SV=1 |
|
| 3.48e-105 | 80 | 785 | 28 | 701 | Beta-galactosidase 11 OS=Oryza sativa subsp. japonica OX=39947 GN=Os08g0549200 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000001 | 0.000023 |