You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_215566T0-p1
You are here: Home > Sequence: PHYCI_215566T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
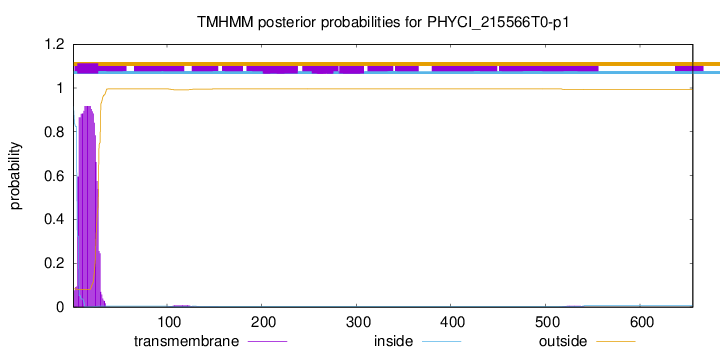
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_215566T0-p1 | |||||||||||
| CAZy Family | PL3|PL3 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 276809 | TPR | 9.57e-20 | 188 | 281 | 1 | 94 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 9.14e-16 | 158 | 250 | 5 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 411346 | social_mot_Tgl | 1.22e-13 | 194 | 279 | 37 | 123 | social motility TPR repeat lipoprotein Tgl. Social motility in delta-proteobacterial species such as Myxococcus xanthus depends on a type VI pilus, which in turn depends on assembly of the PilQ secretin complex. Tgl, a tetratricopeptide repeat (TPR) outer membrane lipoprotein, is required for PilQ assembly. |
| 226428 | Spy | 8.09e-11 | 497 | 652 | 245 | 399 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 223533 | TPR | 2.30e-10 | 169 | 349 | 76 | 261 | Tetratricopeptide (TPR) repeat [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.13e-246 | 18 | 613 | 21 | 622 | |
| 1.02e-24 | 319 | 652 | 366 | 672 | |
| 1.02e-24 | 319 | 652 | 366 | 672 | |
| 1.21e-22 | 310 | 654 | 399 | 712 | |
| 1.88e-22 | 316 | 652 | 93 | 439 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
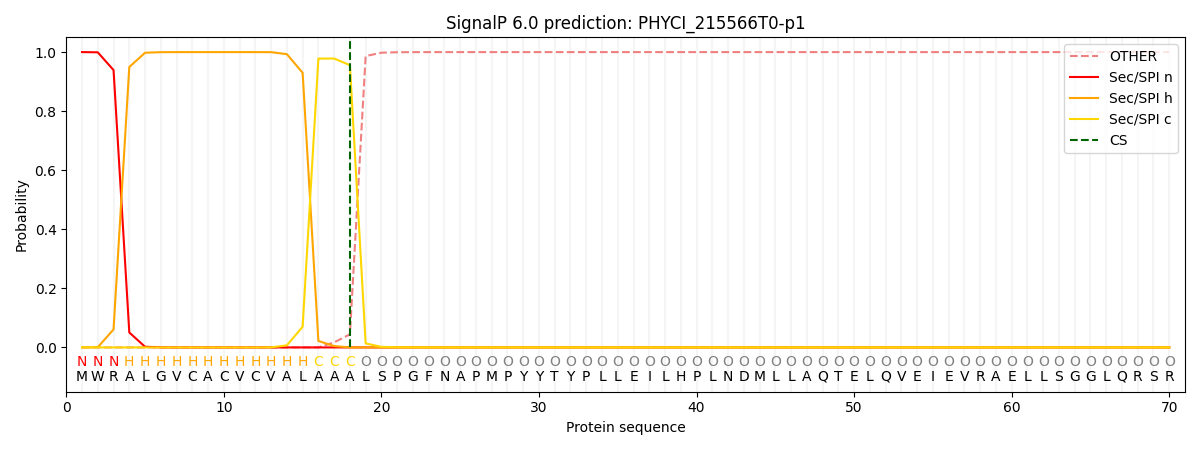
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000232 | 0.999769 | CS pos: 18-19. Pr: 0.9558 |