You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_207198T0-p1
You are here: Home > Sequence: PHYCI_207198T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
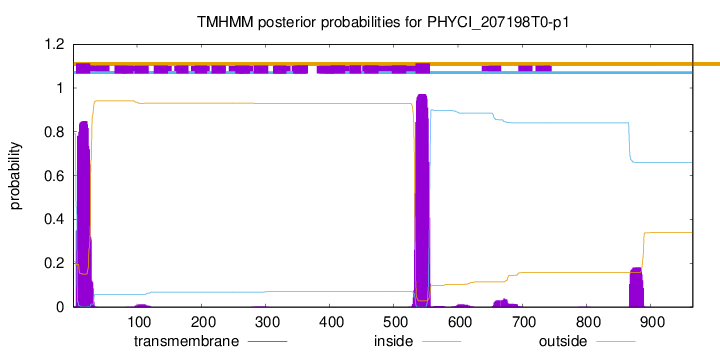
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_207198T0-p1 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH72 | 30 | 378 | 2.6e-73 | 0.967948717948718 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 240334 | PTZ00258 | 0.0 | 578 | 965 | 1 | 387 | GTP-binding protein; Provisional |
| 236583 | PRK09601 | 1.56e-162 | 600 | 965 | 4 | 363 | redox-regulated ATPase YchF. |
| 223091 | GTP1 | 3.33e-150 | 597 | 965 | 1 | 371 | Ribosome-binding ATPase YchF, GTP1/OBG family [Translation, ribosomal structure and biogenesis]. |
| 206687 | YchF | 2.69e-129 | 601 | 880 | 1 | 274 | YchF GTPase. YchF is a member of the Obg family, which includes four other subfamilies of GTPases: Obg, DRG, Ygr210, and NOG1. Obg is an essential gene that is involved in DNA replication in C. crescentus and Streptomyces griseus and is associated with the ribosome. Several members of the family, including YchF, possess the TGS domain related to the RNA-binding proteins. Experimental data and genomic analysis suggest that YchF may be part of a nucleoprotein complex and may function as a GTP-dependent translational factor. |
| 129200 | TIGR00092 | 7.33e-104 | 597 | 965 | 1 | 367 | GTP-binding protein YchF. This predicted GTP-binding protein is found in a single copy in every complete bacterial genome, and is found in Eukaryotes. A more distantly related protein, separated from this model, is found in the archaea. It is known to bind GTP and double-stranded nucleic acid. It is suggested to belong to a nucleoprotein complex and act as a translation factor. [Unknown function, General] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 966 | 1 | 1000 | |
| 0.0 | 31 | 966 | 23 | 953 | |
| 1.16e-214 | 31 | 559 | 332 | 850 | |
| 1.99e-175 | 28 | 470 | 57 | 499 | |
| 2.43e-145 | 31 | 515 | 22 | 500 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.26e-144 | 576 | 965 | 1 | 388 | Crystal structure of OsYchF1 at pH 6.5 [Oryza sativa Japonica Group],5EE1_A Crystal structure of OsYchF1 at pH 7.85 [Oryza sativa Japonica Group],5EE3_A Complex Structure Of Osychf1 With Amp-pnp [Oryza sativa Japonica Group],5EE3_B Complex Structure Of Osychf1 With Amp-pnp [Oryza sativa Japonica Group],5EE9_A Complex structure of OSYCHF1 with GMP-PNP [Oryza sativa Japonica Group],5EE9_B Complex structure of OSYCHF1 with GMP-PNP [Oryza sativa Japonica Group] |
|
| 3.25e-133 | 577 | 965 | 1 | 388 | Crystal structure of human OLA1 in complex with AMPPCP [Homo sapiens] |
|
| 9.51e-122 | 582 | 963 | 4 | 388 | Structure of the Schizosaccharomyces pombe YchF GTPase [Schizosaccharomyces pombe] |
|
| 9.41e-88 | 600 | 965 | 4 | 362 | Ychf Protein (Hi0393) [Haemophilus influenzae],1JAL_B Ychf Protein (Hi0393) [Haemophilus influenzae] |
|
| 1.07e-76 | 601 | 965 | 4 | 367 | Thermus thermophilus YchF GTP-binding protein [Thermus thermophilus HB8],2DWQ_B Thermus thermophilus YchF GTP-binding protein [Thermus thermophilus HB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.16e-144 | 577 | 965 | 1 | 387 | Obg-like ATPase 1 OS=Arabidopsis thaliana OX=3702 GN=YchF1 PE=1 SV=1 |
|
| 2.28e-143 | 577 | 965 | 1 | 387 | Obg-like ATPase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=YCHF1 PE=1 SV=1 |
|
| 1.78e-142 | 577 | 965 | 1 | 387 | Obg-like ATPase 1 OS=Oryza sativa subsp. indica OX=39946 GN=OsI_28170 PE=3 SV=1 |
|
| 7.35e-135 | 577 | 965 | 1 | 386 | Obg-like ATPase 1 OS=Drosophila melanogaster OX=7227 GN=CG1354 PE=1 SV=1 |
|
| 3.92e-134 | 577 | 965 | 1 | 388 | Obg-like ATPase 1 OS=Gallus gallus OX=9031 GN=OLA1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000188 | 0.999773 | CS pos: 24-25. Pr: 0.9601 |