You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_112571T0-p1
You are here: Home > Sequence: PHYCI_112571T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_112571T0-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Vesicle coat complex COPII, subunit SEC23 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.12:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 1063 | 1280 | 4.5e-19 | 0.8883248730964467 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215083 | PLN00162 | 0.0 | 30 | 518 | 227 | 706 | transport protein sec23; Provisional |
| 227380 | SEC23 | 3.26e-177 | 29 | 518 | 221 | 700 | Vesicle coat complex COPII, subunit SEC23 [Intracellular trafficking and secretion]. |
| 238755 | Sec23-like | 3.96e-78 | 29 | 191 | 105 | 267 | Sec23-like: Protein and membrane traffic in eukaryotes is mediated by at least in part by the budding and fusion of intracellular transport vesicles that selectively carry cargo proteins and lipids from donor to acceptor organelles. The two main classes of vesicular carriers within the endocytic and the biosynthetic pathways are COP- and clathrin-coated vesicles. Formation of COPII vesicles requires the ordered assembly of the coat built from several cytosolic components GTPase Sar1, complexes of Sec23-Sec24 and Sec13-Sec31. The process is initiated by the conversion of GDP to GTP by the GTPase Sar1 which then recruits the heterodimeric complex of Sec23 and Sec24. This heterodimeric complex generates the pre-budding complex. The final step leading to membrane deformation and budding of COPII-coated vesicles is carried by the heterodimeric complex Sec13-Sec31. The members of this CD belong to the Sec23-like family. Sec 23 is very similar to Sec24. The Sec23 and Sec24 polypeptides fold into five distinct domains: a beta-barrel, a zinc finger, a vWA or trunk, an all helical region and a carboxy Gelsolin domain. The members of this subgroup lack the consensus MIDAS motif but have the overall Para-Rossmann type fold that is characteristic of this superfamily. |
| 398467 | Sec23_trunk | 7.79e-51 | 29 | 193 | 81 | 241 | Sec23/Sec24 trunk domain. COPII-coated vesicles carry proteins from the endoplasmic reticulum to the Golgi complex. This vesicular transport can be reconstituted by using three cytosolic components containing five proteins: the small GTPase Sar1p, the Sec23p/24p complex, and the Sec13p/Sec31p complex. This domain is known as the trunk domain and has an alpha/beta vWA fold and forms the dimer interface. |
| 200443 | Sec23_C | 8.87e-50 | 422 | 518 | 1 | 97 | C-terminal Actin depolymerization factor-homology domain of Sec23. The C-terminal domain of the Sec23 subunit of the coat protein complex II (COPII) is distantly related to gelsolin-like repeats and the actin depolymerizing domains found in cofilin and similar proteins. Sec23 forms a tight complex with Sec24. The cytoplasmic Sec23/24 complex is recruited together with Sar1-GTP and Sec13/31 to induce coat polymerization and membrane deformation in the forming of COPII-coated endoplasmic reticulum vesicles. The function of the Sec23 C-terminal domain is unclear. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 515 | 1452 | 89 | 1026 | |
| 0.0 | 515 | 1452 | 89 | 1026 | |
| 0.0 | 515 | 1452 | 89 | 1028 | |
| 0.0 | 515 | 1452 | 80 | 1019 | |
| 0.0 | 515 | 1452 | 89 | 1028 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.75e-161 | 24 | 518 | 221 | 706 | Crystal structure of the mammalian COPII-coat protein Sec23/24 bound to the transport signal sequence of membrin [Homo sapiens],3EGD_A Crystal structure of the mammalian COPII-coat protein Sec23a/24a complexed with the SNARE protein Sec22 and bound to the transport signal sequence of vesicular stomatitis virus glycoprotein [Homo sapiens],3EGX_A Crystal structure of the mammalian COPII-coat protein Sec23a/24a complexed with the SNARE protein Sec22b and bound to the transport signal sequence of the SNARE protein Bet1 [Homo sapiens],5VNE_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with Emp24 sorting motif [Homo sapiens],5VNF_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with a C-terminal VV sorting motif [Homo sapiens],5VNG_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with a C-terminal II sorting motif [Homo sapiens],5VNH_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with a C-terminal SV sorting motif [Homo sapiens],5VNI_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with a C-terminal FA sorting motif [Homo sapiens],5VNJ_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with a C-terminal FF sorting motif (ERGIC-53) [Homo sapiens],5VNK_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with a C-terminal LL sorting motif [Homo sapiens],5VNL_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with 4-phenylbutyric acid (1mM soaking) [Homo sapiens],5VNM_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with 4-phenylbutyric acid (15mM soaking) [Homo sapiens],5VNN_A Crystal structure of Sec23a/Sec24a/Sec22 complexed with 4-phenylbutyric acid (50mM soaking) [Homo sapiens],5VNO_A Crystal structure of Sec23a/Sec24a/Sec22 [Homo sapiens] |
|
| 2.83e-161 | 24 | 518 | 221 | 706 | Crystal Structure of the mammalian COPII-coat protein Sec23/24 bound to the transport signal sequence of syntaxin 5 [Homo sapiens] |
|
| 2.83e-161 | 24 | 518 | 221 | 706 | Structure of Sec23 and TANGO1 complex [Homo sapiens],5KYN_B Structure of Sec23 and TANGO1 complex [Homo sapiens],5KYU_A crystal structure of Sec23 and TANGO1 peptide2 complex [Homo sapiens],5KYW_A crystal structure of Sec23 and TANGO1 peptide3 complex [Homo sapiens],5KYX_A crystal structure of Sec23 and TANGO1 peptide1 complex [Homo sapiens],5KYY_A Crystal structure of Sec23 and TANGO1 peptide4 complex [Homo sapiens] |
|
| 3.19e-161 | 24 | 518 | 225 | 710 | Crystal Structure of the human Sec23a/24a heterodimer, complexed with the SNARE protein Sec22b [Homo sapiens],2NUT_A Crystal Structure of the human Sec23a/24a heterodimer, complexed with the SNARE protein Sec22b [Homo sapiens] |
|
| 9.32e-136 | 31 | 518 | 225 | 711 | Cryo-tomography and subtomogram averaging of Sar1-Sec23-Sec24 - fitted model. [Saccharomyces cerevisiae S288C],6ZGA_A Chain A, Protein transport protein SEC23 [Saccharomyces cerevisiae],6ZGA_E Chain E, Protein transport protein SEC23 [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.69e-163 | 1 | 519 | 203 | 709 | Protein transport protein Sec23B OS=Homo sapiens OX=9606 GN=SEC23B PE=1 SV=2 |
|
| 7.34e-163 | 1 | 519 | 203 | 709 | Protein transport protein Sec23B OS=Mus musculus OX=10090 GN=Sec23b PE=1 SV=1 |
|
| 2.80e-162 | 1 | 518 | 203 | 708 | Protein transport protein Sec23B OS=Bos taurus OX=9913 GN=SEC23B PE=2 SV=1 |
|
| 1.07e-161 | 1 | 518 | 203 | 708 | Protein transport protein Sec23A OS=Gallus gallus OX=9031 GN=SEC23A PE=2 SV=1 |
|
| 7.69e-161 | 1 | 519 | 203 | 708 | Protein transport protein Sec23B OS=Pongo abelii OX=9601 GN=SEC23B PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000044 | 0.000001 |
TMHMM Annotations download full data without filtering help
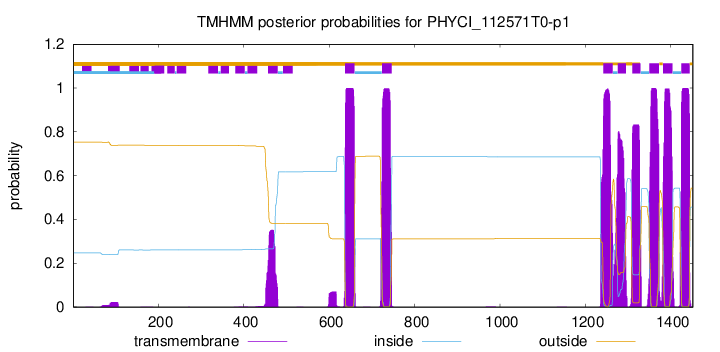
| Start | End |
|---|---|
| 637 | 659 |
| 724 | 746 |
| 1242 | 1264 |
| 1276 | 1295 |
| 1310 | 1329 |
| 1350 | 1372 |
| 1382 | 1404 |
| 1425 | 1444 |
