You are browsing environment: FUNGIDB
PHYCI_103969T0-p1
Basic Information
help
Species
Phytophthora cinnamomi
Lineage
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi
CAZyme ID
PHYCI_103969T0-p1
CAZy Family
AA1
CAZyme Description
unspecified product
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
501
54010.30
4.4185
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_PcinnamomiCBS144-22
26201
1048749
70
26131
Gene Location
Family
Start
End
Evalue
family coverage
GH30
36
491
6.3e-135
0.9868421052631579
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
405300
Glyco_hydr_30_2
3.33e-16
35
240
8
195
O-Glycosyl hydrolase family 30.
more
227807
XynC
2.15e-10
140
491
93
426
O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis].
more
399741
Alpha-L-AF_C
0.005
374
487
59
192
Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
5.60e-227
3
494
4
497
5.83e-165
15
497
21
498
5.17e-161
22
497
29
499
5.17e-161
22
497
29
499
7.32e-161
9
497
16
499
PHYCI_103969T0-p1 has no PDB hit.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5.27e-129
5
498
2
478
Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1
more
3.70e-15
5
368
6
445
Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1
more
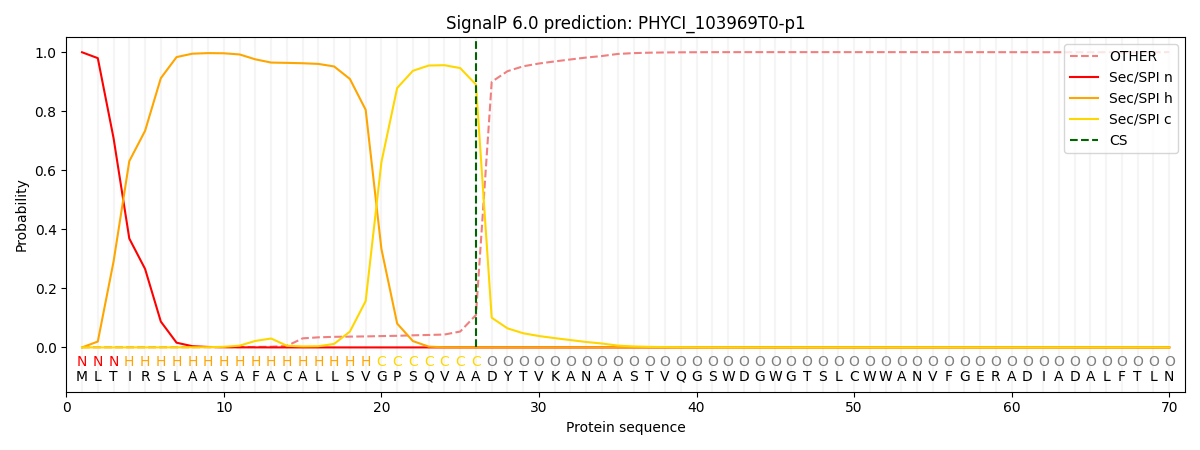
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.001707
0.998239
CS pos: 26-27. Pr: 0.8901
There is no transmembrane helices in PHYCI_103969T0-p1.