You are browsing environment: FUNGIDB
CAZyme Information: PHYCI_103799T0-p1
You are here: Home > Sequence: PHYCI_103799T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | PHYCI_103799T0-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 180051; End:181071 Strand: - | |||||||||||
Full Sequence Download help
| MVASATSFSA LVATAALAAT VSAHGYMSDP AVKFLSSSTD NTQFIATIES SASGFTGSFS | 60 |
| GSPADNTAAF TKAFEASSYS SLKELIDDKA TVTVSDATLT CGSCDPGETD QPLPDTYVEW | 120 |
| AHSDSEGFTS SHEGPCEVWC DDVRVFQDND CAADYTSAPA KLPFDHDACL GTSTLTFYWL | 180 |
| ALHSSTWQVY VNCAPLESTT STGATSKYAV GTSSGTNAST TSNSTSSTTS TTSAPSVTTA | 240 |
| ANTSSSASSA TTSSAASSSY NFQTNYATST SASTSTDTTE TTTAPAATTV TPSVATSTDT | 300 |
| SKCSVRRRRN | 310 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 12 | 275 | 8.4e-71 | 0.9346938775510204 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| EGZ08725.1|AA17 | 4.72e-125 | 1 | 214 | 1 | 213 |
| EGZ08724.1|AA17 | 4.16e-121 | 1 | 214 | 1 | 214 |
| ETI41722.1|AA17 | 4.64e-121 | 1 | 214 | 1 | 214 |
| ETI41723.1|AA17 | 2.11e-120 | 1 | 214 | 1 | 213 |
| ETN20047.1|AA17 | 2.11e-120 | 1 | 214 | 1 | 213 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Z5Y_A | 1.21e-13 | 24 | 196 | 1 | 170 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
SignalP and Lipop Annotations help
This protein is predicted as SP
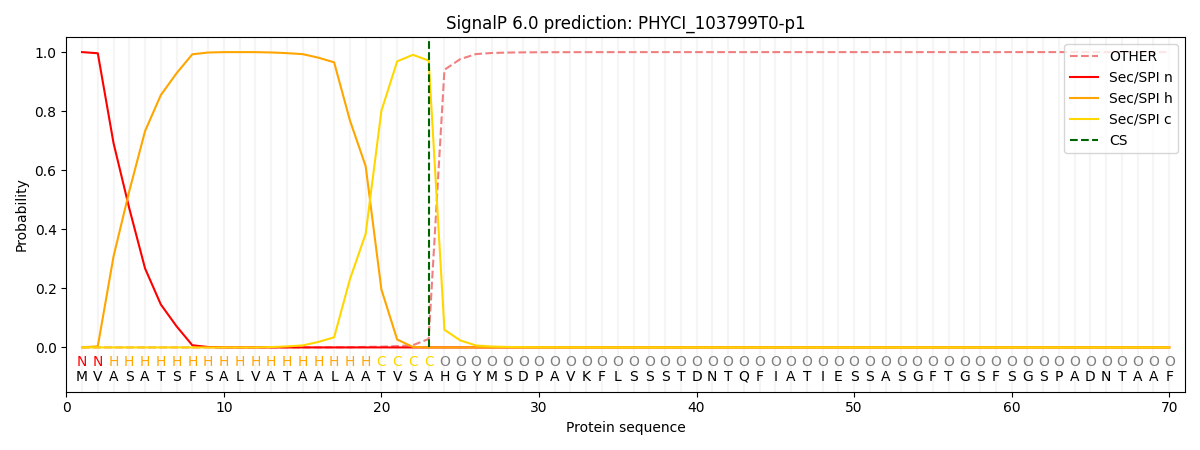
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000410 | 0.999557 | CS pos: 23-24. Pr: 0.9714 |
