You are browsing environment: FUNGIDB
CAZyme Information: PHYBL_79099T0-p1
You are here: Home > Sequence: PHYBL_79099T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phycomyces blakesleeanus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Phycomycetaceae; Phycomyces; Phycomyces blakesleeanus | |||||||||||
| CAZyme ID | PHYBL_79099T0-p1 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL8 | 378 | 595 | 8.8e-39 | 0.8648648648648649 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238517 | GAG_Lyase | 6.46e-69 | 30 | 693 | 9 | 693 | Glycosaminoglycan (GAG) polysaccharide lyase family. This family consists of a group of secreted bacterial lyase enzymes capable of acting on glycosaminoglycans, such as hyaluronan and chondroitin, in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. These are broad-specificity glycosaminoglycan lyases which recognize uronyl residues in polysaccharides and cleave their glycosidic bonds via a beta-elimination reaction to form a double bond between C-4 and C-5 of the non-reducing terminal uronyl residues of released products. Substrates include chondroitin, chondroitin 4-sulfate, chondroitin 6-sulfate, and hyaluronic acid. Family members include chondroitin AC lyase, chondroitin abc lyase, xanthan lyase, and hyalurate lyase. |
| 396729 | Lyase_8 | 4.27e-67 | 377 | 596 | 1 | 222 | Polysaccharide lyase family 8, super-sandwich domain. This family consists of a group of secreted bacterial lyase enzymes EC:4.2.2.1 capable of acting on hyaluronan and chondroitin in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. |
| 400444 | Lyase_8_N | 9.76e-18 | 32 | 298 | 12 | 265 | Polysaccharide lyase family 8, N terminal alpha-helical domain. This family consists of a group of secreted bacterial lyase enzymes EC:4.2.2.1 capable of acting on hyaluronan and chondroitin in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. |
| 397152 | Lyase_8_C | 9.38e-06 | 636 | 700 | 4 | 66 | Polysaccharide lyase family 8, C-terminal beta-sandwich domain. This family consists of a group of secreted bacterial lyase enzymes EC:4.2.2.1 capable of acting on hyaluronan and chondroitin in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. |
| 381256 | globin_sensor | 0.008 | 224 | 272 | 29 | 84 | Globin sensor domain of globin-coupled-sensors (GCSs), protoglobins (Pgbs), and sensor single-domain globins (SSDgbs); S family. This family includes sensor domains which binds porphyrins, and other non-heme cofactors. GCSs have an N-terminal sensor domain coupled to a functional domain. For heme-bound oxygen sensing/binding globin domains, O2 binds to/dissociates from the heme iron complex inducing a structural change in the sensor domain, which is then transduced to the functional domain, switching on (or off) the function of the latter. Functional domains include DGC/GGDEF, EAL, histidine kinase, MCP, PAS, and GAF domains. Characterized members include Bacillus subtilis heme-based aerotaxis transducer (HemAT-Bs) which has a sensor domain coupled to an MCP domain. HemAT-Bs mediates an aerophilic response, and may control the movement direction of bacteria and archaea. Its MCP domain interacts with the CheA histidine kinase, a component of the CheA/CheY signal transduction system that regulates the rotational direction of flagellar motors. Another GCS having the sensor domain coupled to an MCP domain is Caulobacter crescentus McpB. McpB is encoded by a gene which lies adjacent to the major chemotaxis operon. Like McpA (encoded on this operon), McpB has three potential methylation sites, a C-terminal CheBR docking motif, and a motif needed for proteolysis via a ClpX-dependent pathway during the swarmer-to-stalked cell transition. Also included is Geobacter sulfurreducens GCS, a GCS of unknown function, in which the sensor domain is coupled to a transmembrane signal-transduction domain. Pgbs are single-domain globins of unknown function. Methanosarcina acetivorans Pgbs is dimeric and has an N-terminal extension, which together with other Pgb-specific loops, buries the heme within the protein; small ligand molecules gain access to the heme via two orthogonal apolar tunnels. Pgbs and other single-domain globins can function as sensors, when coupled to an appropriate regulator domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.85e-150 | 22 | 706 | 23 | 718 | |
| 1.63e-146 | 42 | 706 | 32 | 732 | |
| 6.42e-146 | 42 | 706 | 32 | 732 | |
| 7.91e-146 | 13 | 706 | 9 | 726 | |
| 8.61e-146 | 19 | 704 | 20 | 731 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.64e-33 | 52 | 585 | 32 | 524 | CHONDROITINASE AC LYASE FROM FLAVOBACTERIUM HEPARINUM [Pedobacter heparinus] |
|
| 1.84e-33 | 52 | 585 | 54 | 546 | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS [Pedobacter heparinus],1HM3_A Active Site Of Chondroitinase Ac Lyase Revealed By The Structure Of Enzyme-Oligosaccharide Complexes And Mutagenesis [Pedobacter heparinus],1HMU_A ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS [Pedobacter heparinus],1HMW_A Active Site Of Chondroitinase Ac Lyase Revealed By The Structure Of Enzyme-oligosaccharide Complexes And Mutagenesis [Pedobacter heparinus] |
|
| 1.63e-18 | 145 | 704 | 122 | 708 | Structure of Paenibacillus xanthan lyase to 2.6 A resolution [Paenibacillus] |
|
| 1.86e-16 | 145 | 591 | 124 | 598 | Crystal structure of the Arthrobacter aurescens chondroitin AC lyase [Paenarthrobacter aurescens] |
|
| 1.86e-16 | 145 | 591 | 124 | 598 | Crystal structure of Arthrobacter aurescens chondroitin AC lyase [Paenarthrobacter aurescens],1RWC_A Crystal structure of Arthrobacter aurescens chondroitin AC lyase [Paenarthrobacter aurescens],1RWF_A Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide [Paenarthrobacter aurescens],1RWG_A Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide [Paenarthrobacter aurescens],1RWH_A Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide [Paenarthrobacter aurescens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.45e-33 | 52 | 585 | 54 | 546 | Chondroitinase-AC OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / CIP 104194 / JCM 7457 / NBRC 12017 / NCIMB 9290 / NRRL B-14731 / HIM 762-3) OX=485917 GN=cslA PE=1 SV=1 |
|
| 8.34e-14 | 198 | 705 | 219 | 757 | Hyaluronate lyase OS=Cutibacterium acnes (strain DSM 16379 / KPA171202) OX=267747 GN=PPA0380 PE=3 SV=1 |
|
| 2.25e-12 | 145 | 533 | 182 | 602 | Hyaluronate lyase OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=hysA PE=3 SV=1 |
|
| 5.68e-12 | 143 | 588 | 368 | 863 | Hyaluronate lyase OS=Streptococcus agalactiae serotype III (strain NEM316) OX=211110 GN=hylB PE=1 SV=2 |
|
| 6.53e-11 | 145 | 646 | 146 | 659 | Xanthan lyase OS=Bacillus sp. (strain GL1) OX=84635 GN=xly PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
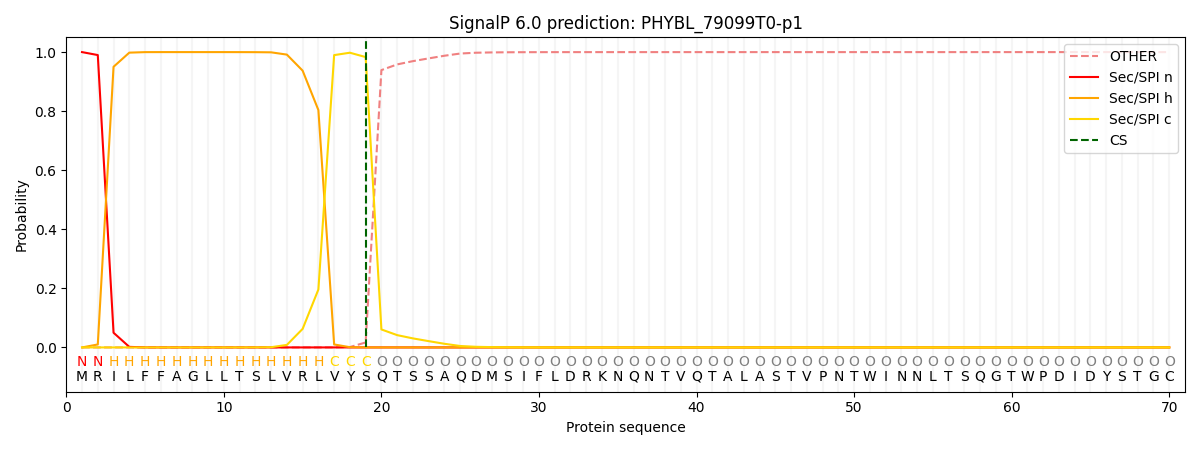
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000228 | 0.999753 | CS pos: 19-20. Pr: 0.9832 |
