You are browsing environment: FUNGIDB
CAZyme Information: PHYBL_76095T0-p1
You are here: Home > Sequence: PHYBL_76095T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
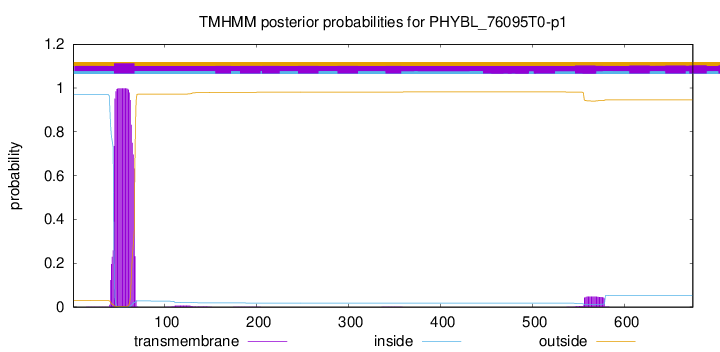
TMHMM annotations
Basic Information help
| Species | Phycomyces blakesleeanus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Phycomycetaceae; Phycomyces; Phycomyces blakesleeanus | |||||||||||
| CAZyme ID | PHYBL_76095T0-p1 | |||||||||||
| CAZy Family | GT34 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT5 | 104 | 632 | 5.7e-51 | 0.9449152542372882 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340822 | GT5_Glycogen_synthase_DULL1-like | 1.70e-35 | 104 | 648 | 2 | 463 | Glycogen synthase GlgA and similar proteins. This family is most closely related to the GT5 family of glycosyltransferases. Glycogen synthase (EC:2.4.1.21) catalyzes the formation and elongation of the alpha-1,4-glucose backbone using ADP-glucose, the second and key step of glycogen biosynthesis. This family includes starch synthases of plants, such as DULL1 in Zea mays and glycogen synthases of various organisms. |
| 223374 | GlgA | 1.08e-22 | 106 | 667 | 5 | 480 | Glycogen synthase [Carbohydrate transport and metabolism]. |
| 234809 | glgA | 8.69e-21 | 106 | 667 | 5 | 465 | glycogen synthase GlgA. |
| 395425 | Glycos_transf_1 | 7.75e-18 | 466 | 616 | 1 | 141 | Glycosyl transferases group 1. Mutations in this domain of PIGA lead to disease (Paroxysmal Nocturnal haemoglobinuria). Members of this family transfer activated sugars to a variety of substrates, including glycogen, Fructose-6-phosphate and lipopolysaccharides. Members of this family transfer UDP, ADP, GDP or CMP linked sugars. The eukaryotic glycogen synthases may be distant members of this family. |
| 404563 | Glyco_trans_1_4 | 3.13e-17 | 467 | 582 | 1 | 112 | Glycosyl transferases group 1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.62e-206 | 26 | 674 | 75 | 810 | |
| 5.62e-188 | 38 | 669 | 63 | 723 | |
| 7.81e-109 | 72 | 673 | 108 | 812 | |
| 2.62e-17 | 268 | 646 | 398 | 776 | |
| 2.62e-17 | 268 | 646 | 398 | 776 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.65e-10 | 446 | 603 | 19 | 175 | Structure of the C domain of glycogen synthase from Pyrococcus abyssi [Pyrococcus abyssi] |
|
| 4.68e-10 | 101 | 584 | 9 | 427 | Catalytic domain of Starch Synthase IV from Arabidopsis thaliana bound to ADP and acarbose [Arabidopsis thaliana],6GNE_B Catalytic domain of Starch Synthase IV from Arabidopsis thaliana bound to ADP and acarbose [Arabidopsis thaliana] |
|
| 1.05e-09 | 446 | 646 | 228 | 433 | Crystal structure of human alpha2C adrenergic G protein-coupled receptor. [Homo sapiens],6KUW_B Crystal structure of human alpha2C adrenergic G protein-coupled receptor. [Homo sapiens] |
|
| 1.07e-09 | 446 | 646 | 220 | 422 | Crystal structure of a class A GPCR [Homo sapiens] |
|
| 1.18e-09 | 446 | 603 | 234 | 390 | Chain A, GlgA glycogen synthase [Pyrococcus abyssi],3L01_B Chain B, GlgA glycogen synthase [Pyrococcus abyssi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.70e-11 | 268 | 585 | 146 | 427 | Glycogen synthase OS=Thermodesulfovibrio yellowstonii (strain ATCC 51303 / DSM 11347 / YP87) OX=289376 GN=glgA PE=3 SV=1 |
|
| 4.66e-11 | 227 | 624 | 103 | 452 | Glycogen synthase 2 OS=Nitrosococcus oceani (strain ATCC 19707 / BCRC 17464 / JCM 30415 / NCIMB 11848 / C-107) OX=323261 GN=glgA2 PE=3 SV=1 |
|
| 1.82e-10 | 374 | 594 | 230 | 415 | Glycogen synthase OS=Sulfurihydrogenibium sp. (strain YO3AOP1) OX=436114 GN=glgA PE=3 SV=1 |
|
| 5.73e-10 | 290 | 584 | 148 | 422 | Glycogen synthase OS=Chlorobaculum parvum (strain DSM 263 / NCIMB 8327) OX=517417 GN=glgA PE=3 SV=1 |
|
| 5.74e-10 | 104 | 624 | 5 | 453 | Glycogen synthase OS=Desulfovibrio vulgaris subsp. vulgaris (strain DP4) OX=391774 GN=glgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
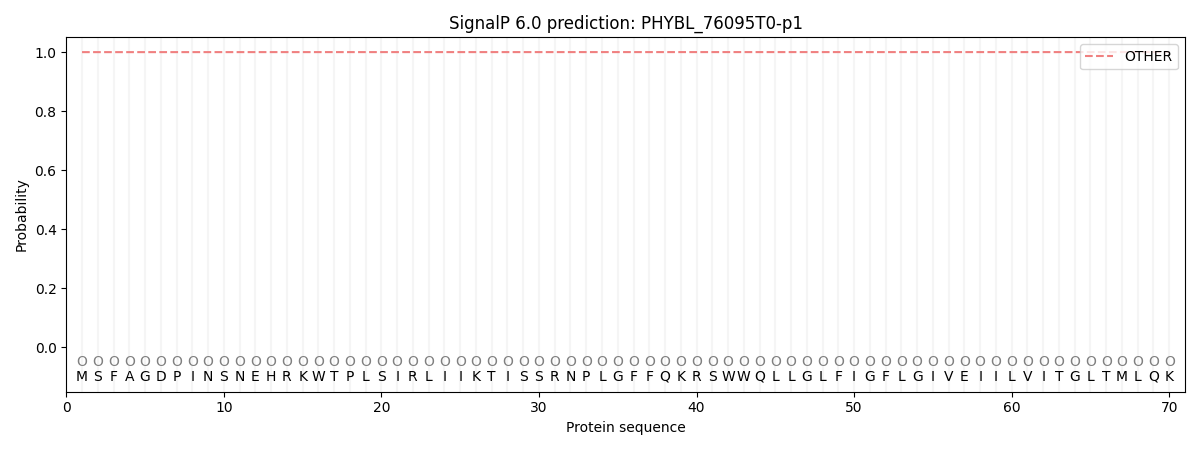
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999981 | 0.000032 |