You are browsing environment: FUNGIDB
CAZyme Information: PHYBL_66143T0-p1
Basic Information
help
| Species |
Phycomyces blakesleeanus
|
| Lineage |
Mucoromycota; Mucoromycetes; ; Phycomycetaceae; Phycomyces; Phycomyces blakesleeanus
|
| CAZyme ID |
PHYBL_66143T0-p1
|
| CAZy Family |
GT20 |
| CAZyme Description |
FKBP-type peptidyl-prolyl cis-trans isomerase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 565 |
|
61761.34 |
4.2244 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PblakesleeanusNRRL1555 |
16528 |
763407 |
0 |
16528
|
|
| Gene Location |
No EC number prediction in PHYBL_66143T0-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| AA12 |
75 |
475 |
4.9e-34 |
0.9825436408977556 |
PHYBL_66143T0-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 4.41e-42 |
88 |
477 |
64 |
461 |
| 1.91e-23 |
149 |
478 |
162 |
508 |
| 1.93e-23 |
149 |
478 |
163 |
509 |
| 1.97e-20 |
70 |
475 |
25 |
453 |
| 3.36e-20 |
79 |
487 |
35 |
466 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 2.73e-11 |
87 |
475 |
16 |
398 |
Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
| 2.74e-11 |
87 |
475 |
17 |
399 |
Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
| 2.75e-11 |
87 |
475 |
18 |
400 |
Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
PHYBL_66143T0-p1 has no Swissprot hit.
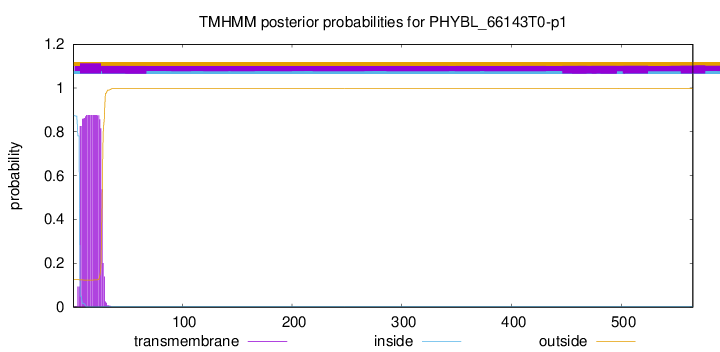
This protein is predicted as SP

| Other |
SP_Sec_SPI |
CS Position |
| 0.000637 |
0.999319 |
CS pos: 26-27. Pr: 0.9733 |