You are browsing environment: FUNGIDB
CAZyme Information: PHYBL_62978T0-p1
You are here: Home > Sequence: PHYBL_62978T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phycomyces blakesleeanus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Phycomycetaceae; Phycomyces; Phycomyces blakesleeanus | |||||||||||
| CAZyme ID | PHYBL_62978T0-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Multicopper oxidases | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 52 | 612 | 3.6e-95 | 0.9776536312849162 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274555 | ascorbase | 2.90e-101 | 33 | 625 | 1 | 530 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 215324 | PLN02604 | 5.75e-87 | 20 | 621 | 15 | 549 | oxidoreductase |
| 177843 | PLN02191 | 1.48e-83 | 33 | 623 | 23 | 551 | L-ascorbate oxidase |
| 132431 | ascorbOXfungal | 3.96e-80 | 38 | 612 | 13 | 525 | L-ascorbate oxidase, fungal type. This model describes a family of fungal ascorbate oxidases, within a larger family of multicopper oxidases that also includes plant ascorbate oxidases (TIGR03388), plant laccases and laccase-like proteins (TIGR03389), and related proteins. The member from Acremonium sp. HI-25 is characterized. |
| 274556 | laccase | 5.75e-57 | 33 | 615 | 3 | 516 | laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.84e-192 | 23 | 623 | 9 | 570 | |
| 2.67e-124 | 33 | 625 | 25 | 647 | |
| 6.69e-123 | 34 | 623 | 28 | 573 | |
| 3.27e-106 | 33 | 591 | 22 | 520 | |
| 3.04e-73 | 6 | 623 | 12 | 551 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.97e-71 | 47 | 628 | 17 | 533 | Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1AOZ_B Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1ASO_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASO_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASP_A X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASP_B X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASQ_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASQ_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo] |
|
| 9.14e-48 | 38 | 622 | 8 | 474 | Type-2 Cu-depleted fungus laccase from Trametes hirsuta [Trametes hirsuta] |
|
| 2.38e-47 | 38 | 622 | 8 | 474 | Native fungus laccase from Trametes hirsuta [Trametes hirsuta],3V9C_A Type-2 Cu-depleted fungus laccase from Trametes hirsuta at low dose of ionization radiation [Trametes hirsuta] |
|
| 1.61e-46 | 55 | 618 | 23 | 469 | Chain A, LACCASE 2 [Trametes versicolor] |
|
| 4.19e-46 | 38 | 622 | 8 | 474 | Recombinant high-redox potential laccase from Basidiomycete Trametes hirsuta [Trametes hirsuta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.78e-73 | 17 | 625 | 14 | 557 | L-ascorbate oxidase OS=Nicotiana tabacum OX=4097 GN=AAO PE=2 SV=1 |
|
| 9.97e-71 | 6 | 628 | 10 | 563 | L-ascorbate oxidase OS=Cucurbita maxima OX=3661 GN=AAO PE=1 SV=2 |
|
| 2.04e-70 | 47 | 628 | 17 | 533 | L-ascorbate oxidase OS=Cucurbita pepo var. melopepo OX=3665 PE=1 SV=1 |
|
| 3.27e-69 | 29 | 623 | 34 | 564 | L-ascorbate oxidase OS=Cucumis sativus OX=3659 PE=1 SV=1 |
|
| 8.75e-66 | 47 | 618 | 34 | 542 | L-ascorbate oxidase OS=Brassica rapa subsp. pekinensis OX=51351 GN=AO PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
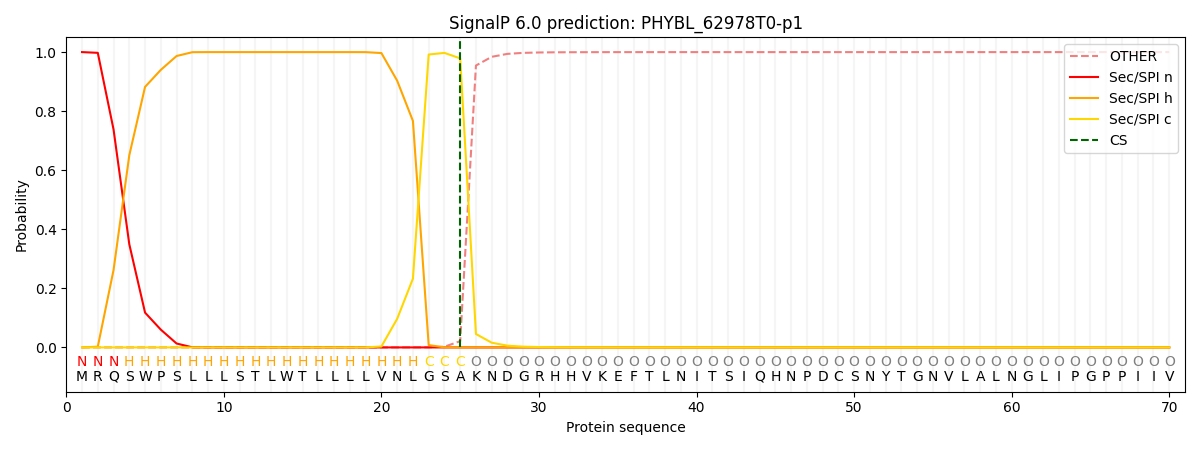
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000198 | 0.999777 | CS pos: 25-26. Pr: 0.9788 |
