You are browsing environment: FUNGIDB
CAZyme Information: PHYBL_185479T0-p1
You are here: Home > Sequence: PHYBL_185479T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phycomyces blakesleeanus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Phycomycetaceae; Phycomyces; Phycomyces blakesleeanus | |||||||||||
| CAZyme ID | PHYBL_185479T0-p1 | |||||||||||
| CAZy Family | GT15 | |||||||||||
| CAZyme Description | Protein involved in dolichol pathway for N-glycosylation (mannosyltransferase family) | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.260:8 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 5 | 388 | 8e-77 | 0.9254498714652957 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 1.91e-35 | 5 | 351 | 2 | 358 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 239511 | GRX_GRXh_1_2_like | 2.15e-35 | 511 | 591 | 2 | 81 | Glutaredoxin (GRX) family, GRX human class 1 and 2 (h_1_2)-like subfamily; composed of proteins similar to human GRXs, approximately 10 kDa in size, and proteins containing a GRX or GRX-like domain. GRX is a glutathione (GSH) dependent reductase, catalyzing the disulfide reduction of target proteins such as ribonucleotide reductase. It contains a redox active CXXC motif in a TRX fold and uses a similar dithiol mechanism employed by TRXs for intramolecular disulfide bond reduction of protein substrates. Unlike TRX, GRX has preference for mixed GSH disulfide substrates, in which it uses a monothiol mechanism where only the N-terminal cysteine is required. The flow of reducing equivalents in the GRX system goes from NADPH -> GSH reductase -> GSH -> GRX -> protein substrates. By altering the redox state of target proteins, GRX is involved in many cellular functions including DNA synthesis, signal transduction and the defense against oxidative stress. Different classes are known including human GRX1 and GRX2, which are members of this subfamily. Also included in this subfamily are the N-terminal GRX domains of proteins similar to human thioredoxin reductase 1 and 3. |
| 274016 | GRX_euk | 3.32e-34 | 511 | 593 | 1 | 83 | Glutaredoxin. Glutaredoxins are thioltransferases (disulfide reductases) which utilize glutathione and NADPH as cofactors. Oxidized glutathione is regenerated by glutathione reductase. Together these components compose the glutathione system. Glutaredoxins utilize the CXXC motif common to thioredoxins and are involved in multiple cellular processes including protection from redox stress, reduction of critical enzymes such as ribonucleotide reductase and the generation of reduced sulfur for iron sulfur cluster formation. Glutaredoxins are capable of reduction of mixed disulfides of glutathione as well as the formation of glutathione mixed disulfides. This model represents eukaryotic glutaredoxins and includes sequences from fungi, plants and metazoans as well as viruses. |
| 239017 | GRX_family | 1.80e-25 | 511 | 584 | 2 | 71 | Glutaredoxin (GRX) family; composed of GRX, approximately 10 kDa in size, and proteins containing a GRX or GRX-like domain. GRX is a glutathione (GSH) dependent reductase, catalyzing the disulfide reduction of target proteins such as ribonucleotide reductase. It contains a redox active CXXC motif in a TRX fold and uses a similar dithiol mechanism employed by TRXs for intramolecular disulfide bond reduction of protein substrates. Unlike TRX, GRX has preference for mixed GSH disulfide substrates, in which it uses a monothiol mechanism where only the N-terminal cysteine is required. The flow of reducing equivalents in the GRX system goes from NADPH -> GSH reductase -> GSH -> GRX -> protein substrates. By altering the redox state of target proteins, GRX is involved in many cellular functions including DNA synthesis, signal transduction and the defense against oxidative stress. Different classes are known including human GRX1 and GRX2, as well as E. coli GRX1 and GRX3, which are members of this family. E. coli GRX2, however, is a 24-kDa protein that belongs to the GSH S-transferase (GST) family. |
| 223767 | GrxC | 1.35e-20 | 509 | 590 | 1 | 80 | Glutaredoxin [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.19e-205 | 10 | 595 | 9 | 592 | |
| 1.78e-85 | 12 | 497 | 11 | 479 | |
| 2.81e-84 | 17 | 497 | 16 | 479 | |
| 2.99e-83 | 17 | 497 | 16 | 479 | |
| 6.21e-83 | 17 | 497 | 16 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.58e-21 | 509 | 593 | 17 | 101 | Chain A, Glutaredoxin, CPYC type [Chlamydomonas reinhardtii],7NCW_A Chain A, Glutaredoxin, CPYC type [Chlamydomonas reinhardtii] |
|
| 6.24e-19 | 499 | 593 | 5 | 100 | Crystal structure of the oxidized glutaredoxin from Chlorella sorokiniana T-89 [Chlorella sorokiniana],4I2U_A Crystal structure of the reduced glutaredoxin from Chlorella sorokiniana T-89 in complex with glutathione [Chlorella sorokiniana] |
|
| 1.82e-18 | 506 | 598 | 44 | 139 | Zebrafish Grx2 (APO) [Danio rerio],3UIW_B Zebrafish Grx2 (APO) [Danio rerio] |
|
| 6.06e-17 | 511 | 589 | 65 | 142 | Crystal Structure of Wheat Glutarredoxin [Triticum aestivum],5ZVL_B Crystal Structure of Wheat Glutarredoxin [Triticum aestivum],5ZVL_C Crystal Structure of Wheat Glutarredoxin [Triticum aestivum],5ZVL_D Crystal Structure of Wheat Glutarredoxin [Triticum aestivum],5ZVL_E Crystal Structure of Wheat Glutarredoxin [Triticum aestivum] |
|
| 3.70e-16 | 506 | 593 | 34 | 120 | Crystal structure of buckwheat glutaredoxin-glutathione complex [Polygonaceae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.59e-74 | 8 | 493 | 19 | 482 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Homo sapiens OX=9606 GN=ALG12 PE=1 SV=1 |
|
| 4.52e-74 | 11 | 433 | 25 | 442 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Arabidopsis thaliana OX=3702 GN=ALG12 PE=1 SV=1 |
|
| 1.08e-70 | 12 | 493 | 24 | 480 | Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Mus musculus OX=10090 GN=Alg12 PE=2 SV=2 |
|
| 2.67e-70 | 4 | 493 | 3 | 476 | Probable Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Drosophila melanogaster OX=7227 GN=Alg12 PE=2 SV=1 |
|
| 1.36e-68 | 12 | 420 | 14 | 422 | Probable Dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase OS=Caenorhabditis elegans OX=6239 GN=algn-12 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
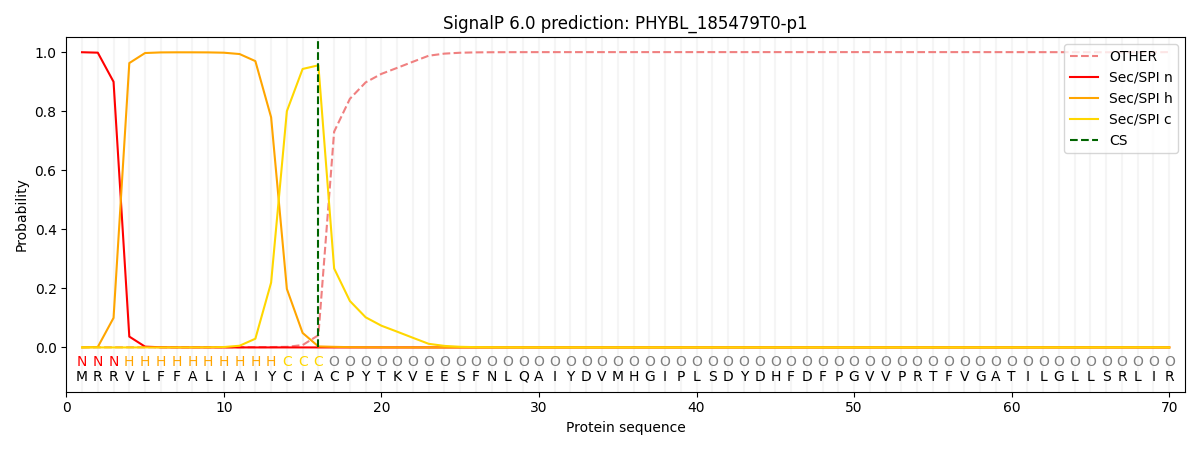
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.002535 | 0.997445 | CS pos: 16-17. Pr: 0.9553 |
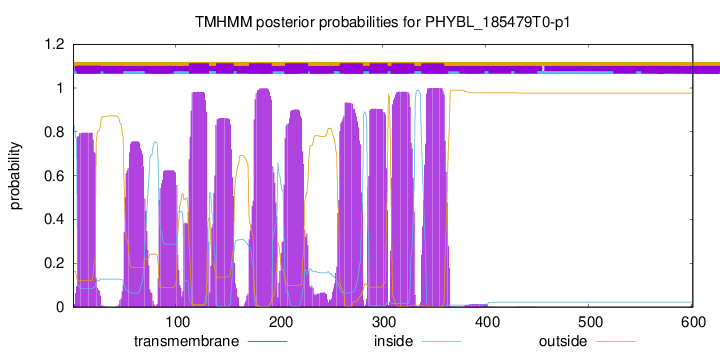
TMHMM Annotations download full data without filtering help
| Start | End |
|---|---|
| 113 | 132 |
| 139 | 156 |
| 171 | 193 |
| 206 | 228 |
| 259 | 281 |
| 288 | 305 |
| 309 | 331 |
| 338 | 360 |
