You are browsing environment: FUNGIDB
CAZyme Information: PHYBL_179151T0-p1
You are here: Home > Sequence: PHYBL_179151T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phycomyces blakesleeanus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Phycomycetaceae; Phycomyces; Phycomyces blakesleeanus | |||||||||||
| CAZyme ID | PHYBL_179151T0-p1 | |||||||||||
| CAZy Family | GH72 | |||||||||||
| CAZyme Description | Mannosyl-oligosaccharide alpha-1,2-mannosidase and related glycosyl hydrolases | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.113:73 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 69 | 526 | 2.6e-144 | 0.9977578475336323 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396217 | Glyco_hydro_47 | 0.0 | 69 | 526 | 1 | 453 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| 240427 | PTZ00470 | 1.32e-155 | 58 | 529 | 68 | 521 | glycoside hydrolase family 47 protein; Provisional |
| 224250 | YyaL | 0.005 | 151 | 262 | 472 | 583 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.51e-111 | 62 | 529 | 34 | 486 | |
| 1.04e-104 | 52 | 524 | 94 | 563 | |
| 3.26e-101 | 66 | 526 | 57 | 529 | |
| 4.29e-99 | 60 | 527 | 180 | 629 | |
| 9.62e-99 | 52 | 542 | 136 | 601 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.54e-91 | 58 | 536 | 16 | 474 | Structure of mouse Golgi alpha-1,2-mannosidase IA reveals the molecular basis for substrate specificity among Class I enzymes (family 47 glycosidases) [Mus musculus] |
|
| 1.31e-90 | 58 | 527 | 18 | 468 | Structure of mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex [Mus musculus],5KKB_B Structure of mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex [Mus musculus] |
|
| 2.59e-88 | 58 | 526 | 1 | 473 | Penicillium citrinum alpha-1,2-mannosidase complex with glycerol [Penicillium citrinum],2RI8_B Penicillium citrinum alpha-1,2-mannosidase complex with glycerol [Penicillium citrinum],2RI9_A Penicillium citrinum alpha-1,2-mannosidase in complex with a substrate analog [Penicillium citrinum],2RI9_B Penicillium citrinum alpha-1,2-mannosidase in complex with a substrate analog [Penicillium citrinum] |
|
| 6.99e-88 | 58 | 526 | 36 | 508 | Structure of P. citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the ER and Golgi Class I enzymes [Penicillium citrinum],1KKT_B Structure of P. citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the ER and Golgi Class I enzymes [Penicillium citrinum],1KRE_A Structure Of P. Citrinum Alpha 1,2-Mannosidase Reveals The Basis For Differences In Specificity Of The Er And Golgi Class I Enzymes [Penicillium citrinum],1KRE_B Structure Of P. Citrinum Alpha 1,2-Mannosidase Reveals The Basis For Differences In Specificity Of The Er And Golgi Class I Enzymes [Penicillium citrinum],1KRF_A Structure Of P. Citrinum Alpha 1,2-mannosidase Reveals The Basis For Differences In Specificity Of The Er And Golgi Class I Enzymes [Penicillium citrinum],1KRF_B Structure Of P. Citrinum Alpha 1,2-mannosidase Reveals The Basis For Differences In Specificity Of The Er And Golgi Class I Enzymes [Penicillium citrinum] |
|
| 1.34e-84 | 62 | 526 | 5 | 451 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.73e-95 | 50 | 526 | 30 | 510 | Mannosyl-oligosaccharide alpha-1,2-mannosidase 1B OS=Aspergillus phoenicis OX=5063 GN=mns1B PE=2 SV=1 |
|
| 3.81e-95 | 60 | 527 | 178 | 627 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB OS=Homo sapiens OX=9606 GN=MAN1A2 PE=1 SV=1 |
|
| 7.52e-95 | 53 | 527 | 171 | 627 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB OS=Mus musculus OX=10090 GN=Man1a2 PE=1 SV=1 |
|
| 3.53e-94 | 9 | 526 | 2 | 514 | Mannosyl-oligosaccharide alpha-1,2-mannosidase OS=Coccidioides posadasii (strain RMSCC 757 / Silveira) OX=443226 GN=CPSG_02648 PE=1 SV=1 |
|
| 5.29e-94 | 57 | 526 | 35 | 507 | Probable mannosyl-oligosaccharide alpha-1,2-mannosidase ARB_00035 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_00035 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
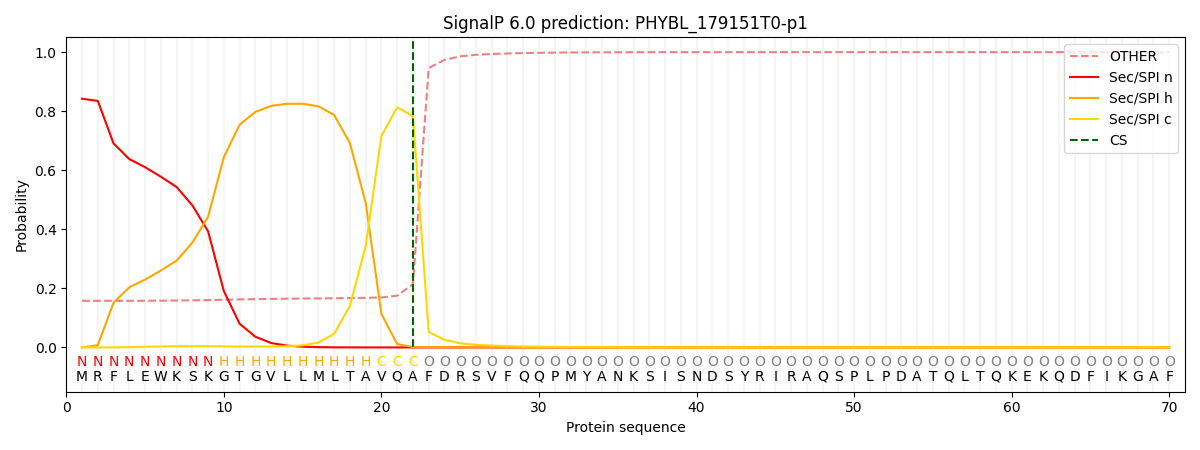
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.167572 | 0.832406 | CS pos: 22-23. Pr: 0.7839 |

