You are browsing environment: FUNGIDB
CAZyme Information: PHYBL_116472T0-p1
You are here: Home > Sequence: PHYBL_116472T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phycomyces blakesleeanus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Phycomycetaceae; Phycomyces; Phycomyces blakesleeanus | |||||||||||
| CAZyme ID | PHYBL_116472T0-p1 | |||||||||||
| CAZy Family | CE20 | |||||||||||
| CAZyme Description | Mannosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.259:12 | 2.4.1.261:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 2 | 470 | 7.3e-97 | 0.9897172236503856 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 6.17e-92 | 2 | 470 | 5 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 215437 | PLN02816 | 1.29e-05 | 235 | 442 | 256 | 426 | mannosyltransferase |
| 224720 | ArnT | 0.010 | 2 | 372 | 16 | 346 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.66e-244 | 73 | 609 | 1 | 537 | |
| 3.86e-152 | 1 | 610 | 64 | 673 | |
| 7.57e-148 | 1 | 610 | 55 | 690 | |
| 9.30e-144 | 1 | 609 | 70 | 687 | |
| 1.55e-141 | 4 | 609 | 37 | 587 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.44e-124 | 2 | 609 | 20 | 560 | Alpha-1,2-mannosyltransferase alg9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg9 PE=3 SV=1 |
|
| 4.95e-116 | 3 | 610 | 65 | 601 | Alpha-1,2-mannosyltransferase ALG9 OS=Mus musculus OX=10090 GN=Alg9 PE=2 SV=1 |
|
| 1.08e-114 | 3 | 610 | 65 | 601 | Alpha-1,2-mannosyltransferase ALG9 OS=Homo sapiens OX=9606 GN=ALG9 PE=1 SV=2 |
|
| 1.35e-87 | 1 | 593 | 58 | 583 | Alpha-1,2-mannosyltransferase algn-9 OS=Caenorhabditis elegans OX=6239 GN=algn-9 PE=1 SV=2 |
|
| 4.17e-75 | 16 | 608 | 28 | 549 | Alpha-1,2-mannosyltransferase ALG9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ALG9 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.934450 | 0.065549 |
TMHMM Annotations download full data without filtering help
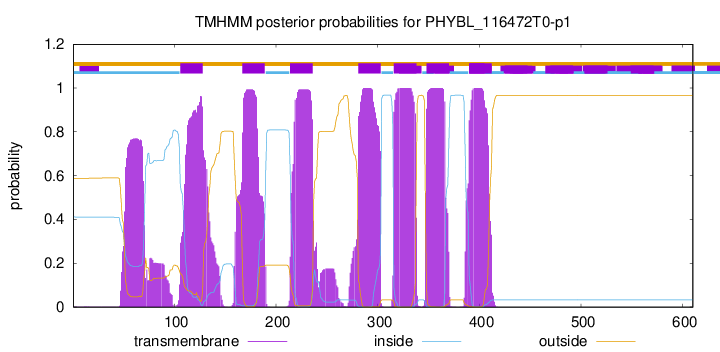
| Start | End |
|---|---|
| 106 | 128 |
| 167 | 189 |
| 214 | 236 |
| 281 | 303 |
| 316 | 338 |
| 348 | 370 |
| 390 | 412 |
