You are browsing environment: FUNGIDB
CAZyme Information: PHPALM_5033-t46_1-p1
You are here: Home > Sequence: PHPALM_5033-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
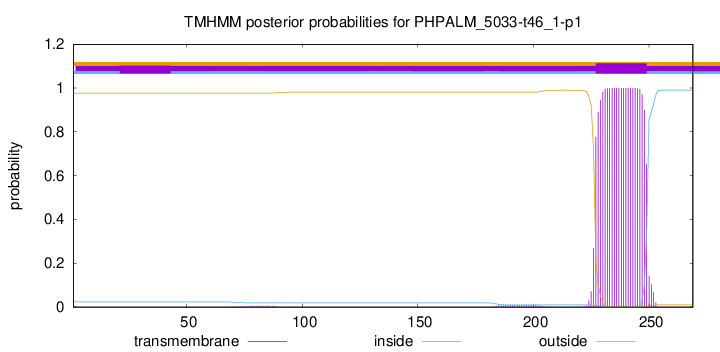
TMHMM annotations
Basic Information help
| Species | Phytophthora palmivora | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora palmivora | |||||||||||
| CAZyme ID | PHPALM_5033-t46_1-p1 | |||||||||||
| CAZy Family | GT31|GT10 | |||||||||||
| CAZyme Description | Glucan 1,3-beta-glucosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM43 | 135 | 206 | 2e-16 | 0.9036144578313253 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 197867 | X8 | 5.38e-16 | 135 | 207 | 3 | 79 | Possibly involved in carbohydrate binding. The X8 domain, which may be involved in carbohydrate binding, is found in an Olive pollen antigen as well as at the C terminus of family 17 glycosyl hydrolases. It contains 6 conserved cysteine residues which presumably form three disulfide bridges. |
| 400371 | X8 | 4.79e-14 | 135 | 200 | 3 | 76 | X8 domain. The X8 domain domain contains at least 6 conserved cysteine residues that presumably form three disulphide bridges. The domain is found in an Olive pollen allergen as well as at the C-terminus of several families of glycosyl hydrolases. This domain may be involved in carbohydrate binding. This domain is characteristic of GPI-anchored domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.07e-157 | 1 | 269 | 402 | 673 | |
| 9.51e-124 | 1 | 269 | 424 | 692 | |
| 2.76e-122 | 1 | 269 | 297 | 566 | |
| 1.34e-120 | 1 | 265 | 422 | 684 | |
| 1.19e-113 | 1 | 264 | 415 | 676 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.80e-07 | 107 | 208 | 335 | 439 | Glucan endo-1,3-beta-glucosidase 4 OS=Arabidopsis thaliana OX=3702 GN=At3g13560 PE=2 SV=1 |
|
| 6.89e-07 | 135 | 210 | 22 | 97 | PLASMODESMATA CALLOSE-BINDING PROTEIN 4 OS=Arabidopsis thaliana OX=3702 GN=PDCB4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000048 | 0.000000 |