You are browsing environment: FUNGIDB
CAZyme Information: PHPALM_37770-t46_1-p1
You are here: Home > Sequence: PHPALM_37770-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
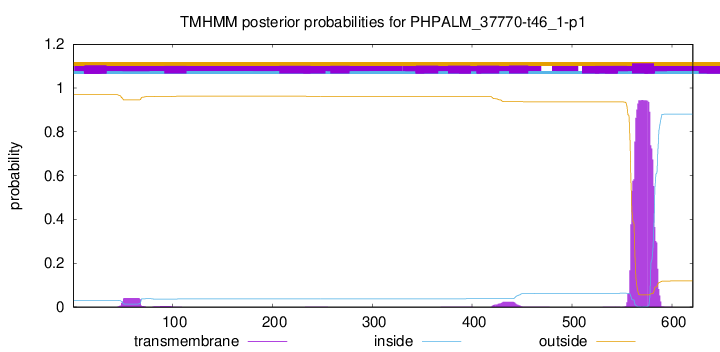
TMHMM annotations
Basic Information help
| Species | Phytophthora palmivora | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora palmivora | |||||||||||
| CAZyme ID | PHPALM_37770-t46_1-p1 | |||||||||||
| CAZy Family | GH81 | |||||||||||
| CAZyme Description | Hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.276:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 176 | 539 | 1.9e-22 | 0.7827225130890052 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 2.46e-33 | 91 | 535 | 3 | 401 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 224732 | YjiC | 8.34e-26 | 90 | 540 | 3 | 400 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 278624 | UDPGT | 9.65e-08 | 231 | 581 | 163 | 482 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 223071 | egt | 2.04e-05 | 382 | 518 | 315 | 442 | ecdysteroid UDP-glucosyltransferase; Provisional |
| 340818 | GT28_MurG | 7.64e-04 | 453 | 519 | 267 | 339 | undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase. MurG (EC 2.4.1.227) is an N-acetylglucosaminyltransferase, the last enzyme involved in the intracellular phase of peptidoglycan biosynthesis. It transfers N-acetyl-D-glucosamine (GlcNAc) from UDP-GlcNAc to the C4 hydroxyl of a lipid-linked N-acetylmuramoyl pentapeptide (NAM). The resulting disaccharide is then transported across the cell membrane, where it is polymerized into NAG-NAM cell-wall repeat structure. MurG belongs to the GT-B structural superfamily of glycoslytransferases, which have characteristic N- and C-terminal domains, each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 6 | 590 | 7 | 592 | |
| 3.24e-43 | 90 | 579 | 89 | 599 | |
| 7.92e-42 | 91 | 579 | 66 | 535 | |
| 3.70e-27 | 72 | 567 | 18 | 508 | |
| 7.48e-22 | 102 | 545 | 1 | 420 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.68e-08 | 90 | 537 | 5 | 383 | Chain A, Uncharacterized UDP-glucosyltransferase YjiC [Bacillus subtilis subsp. subtilis str. 168] |
|
| 2.72e-08 | 90 | 537 | 5 | 383 | Chain A, Uncharacterized UDP-glucosyltransferase YjiC [Bacillus subtilis subsp. subtilis str. 168],7BOV_A Chain A, Uncharacterized UDP-glucosyltransferase YjiC [Bacillus subtilis subsp. subtilis str. 168] |
|
| 4.71e-06 | 335 | 544 | 236 | 433 | Structural Characterization of UDP-glycosyltransferase from Tetranychus Urticae [Tetranychus urticae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.24e-13 | 185 | 571 | 122 | 487 | 2-hydroxyacylsphingosine 1-beta-galactosyltransferase OS=Mus musculus OX=10090 GN=Ugt8 PE=1 SV=2 |
|
| 3.91e-13 | 185 | 577 | 122 | 488 | 2-hydroxyacylsphingosine 1-beta-galactosyltransferase OS=Homo sapiens OX=9606 GN=UGT8 PE=1 SV=2 |
|
| 3.91e-13 | 106 | 570 | 39 | 486 | 2-hydroxyacylsphingosine 1-beta-galactosyltransferase OS=Rattus norvegicus OX=10116 GN=Ugt8 PE=1 SV=1 |
|
| 4.96e-13 | 163 | 564 | 105 | 489 | UDP-glucuronosyltransferase 3A2 OS=Homo sapiens OX=9606 GN=UGT3A2 PE=2 SV=1 |
|
| 4.67e-12 | 186 | 573 | 130 | 498 | UDP-glucuronosyltransferase 3A1 OS=Bos taurus OX=9913 GN=UGT3A1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999692 | 0.000301 |