You are browsing environment: FUNGIDB
CAZyme Information: PHPALM_28146-t46_1-p1
You are here: Home > Sequence: PHPALM_28146-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
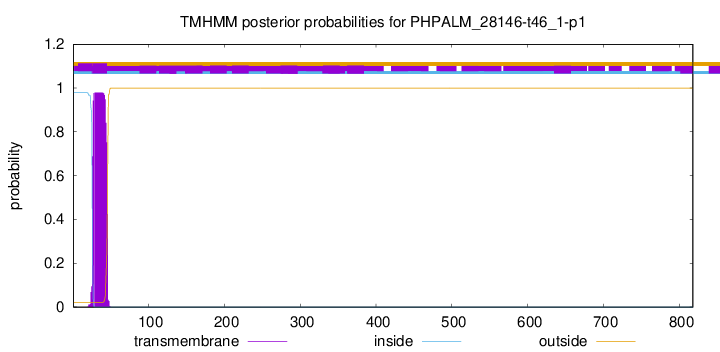
TMHMM annotations
Basic Information help
| Species | Phytophthora palmivora | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora palmivora | |||||||||||
| CAZyme ID | PHPALM_28146-t46_1-p1 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 88 | 389 | 1.3e-86 | 0.993485342019544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 166698 | PLN03059 | 1.40e-122 | 80 | 787 | 29 | 728 | beta-galactosidase; Provisional |
| 396048 | Glyco_hydro_35 | 1.38e-96 | 88 | 388 | 2 | 315 | Glycosyl hydrolases family 35. |
| 224786 | GanA | 3.39e-16 | 81 | 252 | 1 | 173 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 5 | 799 | 9 | 805 | |
| 1.14e-243 | 11 | 787 | 28 | 794 | |
| 5.61e-159 | 61 | 776 | 17 | 721 | |
| 1.66e-154 | 65 | 786 | 22 | 735 | |
| 2.34e-154 | 61 | 776 | 17 | 721 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.04e-105 | 76 | 783 | 3 | 703 | Chain A, Beta-galactosidase [Solanum lycopersicum],3W5F_B Chain B, Beta-galactosidase [Solanum lycopersicum],3W5G_A Chain A, Beta-galactosidase [Solanum lycopersicum],3W5G_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK5_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK5_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
|
| 7.80e-105 | 76 | 783 | 3 | 703 | Chain A, Beta-galactosidase [Solanum lycopersicum],6IK6_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK7_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK7_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK8_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK8_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
|
| 1.62e-41 | 88 | 399 | 24 | 347 | Chain A, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
|
| 2.82e-33 | 78 | 387 | 17 | 345 | Galactanase BT0290 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 4.10e-33 | 88 | 353 | 10 | 284 | Crystal structure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8C_B Crystal structure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8D_A Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4],4E8D_B Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.61e-113 | 80 | 787 | 30 | 731 | Beta-galactosidase 5 OS=Arabidopsis thaliana OX=3702 GN=BGAL5 PE=2 SV=1 |
|
| 2.32e-107 | 80 | 787 | 26 | 726 | Beta-galactosidase 5 OS=Oryza sativa subsp. japonica OX=39947 GN=Os03g0165400 PE=2 SV=1 |
|
| 1.71e-106 | 81 | 787 | 33 | 733 | Beta-galactosidase 3 OS=Arabidopsis thaliana OX=3702 GN=BGAL3 PE=2 SV=1 |
|
| 9.93e-106 | 80 | 787 | 30 | 730 | Putative beta-galactosidase OS=Dianthus caryophyllus OX=3570 GN=CARSR12 PE=2 SV=1 |
|
| 3.14e-105 | 80 | 784 | 31 | 734 | Beta-galactosidase 8 OS=Arabidopsis thaliana OX=3702 GN=BGAL8 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000017 | 0.000013 |