You are browsing environment: FUNGIDB
CAZyme Information: PHPALM_12364-t46_1-p1
You are here: Home > Sequence: PHPALM_12364-t46_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
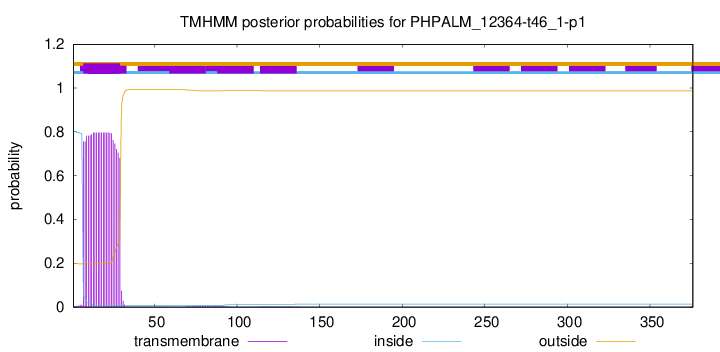
TMHMM annotations
Basic Information help
| Species | Phytophthora palmivora | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora palmivora | |||||||||||
| CAZyme ID | PHPALM_12364-t46_1-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Glycoside hydrolase, family 5 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.78:7 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 55 | 340 | 3.3e-84 | 0.9930795847750865 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226444 | COG3934 | 1.54e-32 | 126 | 373 | 83 | 316 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 1.80e-09 | 147 | 338 | 88 | 266 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.17e-106 | 19 | 332 | 97 | 404 | |
| 4.59e-104 | 19 | 361 | 90 | 431 | |
| 3.89e-103 | 8 | 376 | 13 | 379 | |
| 1.31e-101 | 19 | 375 | 100 | 459 | |
| 1.59e-101 | 19 | 375 | 91 | 454 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.86e-84 | 18 | 376 | 2 | 385 | The Crystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase [Neurospora sitophila] |
|
| 8.42e-81 | 19 | 328 | 1 | 313 | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
|
| 1.05e-71 | 19 | 341 | 1 | 309 | Chain A, ENDO-1,4-B-D-MANNANASE [Trichoderma reesei],1QNP_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNQ_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNR_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNS_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei] |
|
| 3.52e-70 | 32 | 356 | 22 | 361 | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
|
| 4.74e-70 | 21 | 359 | 1 | 330 | The ligand-free structure of ManBK from Aspergillus niger BK01 [Aspergillus niger],3WH9_B The ligand-free structure of ManBK from Aspergillus niger BK01 [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.34e-84 | 10 | 376 | 6 | 396 | Mannan endo-1,4-beta-mannosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manC PE=1 SV=1 |
|
| 6.17e-83 | 18 | 376 | 24 | 403 | Probable mannan endo-1,4-beta-mannosidase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=manC PE=3 SV=1 |
|
| 2.61e-78 | 17 | 365 | 22 | 358 | Mannan endo-1,4-beta-mannosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manD PE=3 SV=1 |
|
| 4.23e-77 | 19 | 355 | 111 | 439 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manF PE=3 SV=1 |
|
| 1.03e-76 | 17 | 341 | 27 | 339 | Mannan endo-1,4-beta-mannosidase A OS=Aspergillus aculeatus OX=5053 GN=manA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000287 | 0.999692 | CS pos: 20-21. Pr: 0.9738 |