You are browsing environment: FUNGIDB
CAZyme Information: PGTG_15158-t26_1-p1
You are here: Home > Sequence: PGTG_15158-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia graminis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia graminis | |||||||||||
| CAZyme ID | PGTG_15158-t26_1-p1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.78:1 |
|---|
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.01e-85 | 139 | 442 | 5 | 340 | |
| 1.67e-82 | 76 | 495 | 57 | 479 | |
| 1.54e-47 | 83 | 484 | 38 | 410 | |
| 6.00e-46 | 295 | 616 | 1 | 319 | |
| 1.70e-41 | 73 | 484 | 31 | 431 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.09e-11 | 78 | 310 | 14 | 232 | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
|
| 6.53e-09 | 161 | 299 | 102 | 223 | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
|
| 1.05e-08 | 161 | 299 | 88 | 209 | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.56e-10 | 75 | 382 | 46 | 338 | Mannan endo-1,4-beta-mannosidase 5 OS=Solanum lycopersicum OX=4081 GN=MAN5 PE=2 SV=1 |
|
| 9.56e-10 | 75 | 382 | 46 | 338 | Mannan endo-1,4-beta-mannosidase 2 OS=Solanum lycopersicum OX=4081 GN=MAN2 PE=2 SV=2 |
|
| 9.63e-09 | 78 | 304 | 41 | 246 | Mannan endo-1,4-beta-mannosidase 7 OS=Arabidopsis thaliana OX=3702 GN=MAN7 PE=2 SV=1 |
|
| 2.56e-08 | 151 | 300 | 134 | 285 | Putative mannan endo-1,4-beta-mannosidase 5 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN5 PE=2 SV=2 |
|
| 9.11e-08 | 152 | 290 | 110 | 236 | Mannan endo-1,4-beta-mannosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN1 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
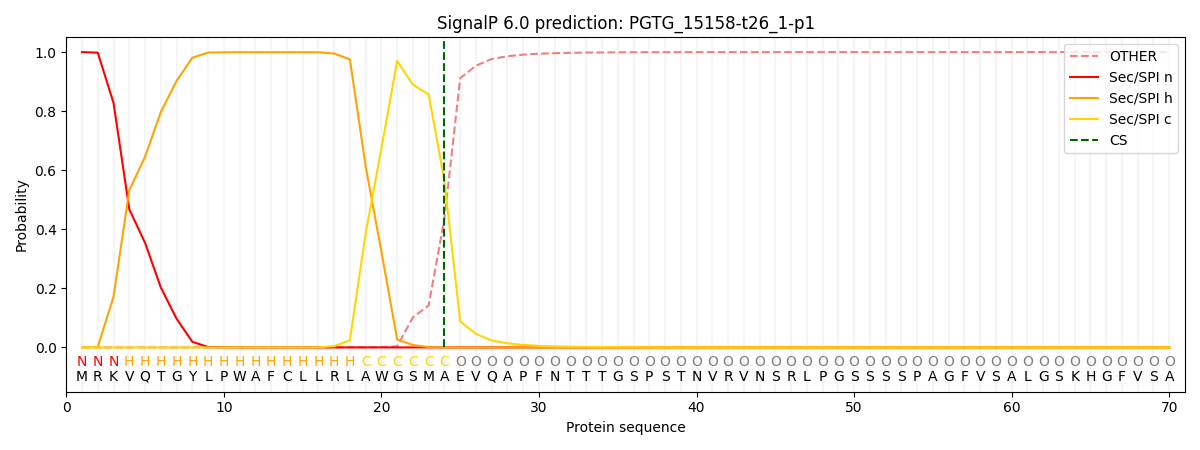
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000463 | 0.999524 | CS pos: 24-25. Pr: 0.5633 |
