You are browsing environment: FUNGIDB
CAZyme Information: PGTG_11088-t26_1-p1
You are here: Home > Sequence: PGTG_11088-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia graminis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia graminis | |||||||||||
| CAZyme ID | PGTG_11088-t26_1-p1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.78:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 78 | 421 | 2.2e-83 | 0.9826989619377162 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226444 | COG3934 | 1.26e-13 | 74 | 471 | 79 | 465 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| 226444 | COG3934 | 6.71e-10 | 53 | 390 | 12 | 268 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 0.005 | 223 | 365 | 86 | 237 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.47e-266 | 6 | 474 | 8 | 476 | |
| 1.83e-151 | 36 | 478 | 31 | 476 | |
| 1.53e-148 | 37 | 486 | 32 | 491 | |
| 1.13e-144 | 12 | 475 | 10 | 488 | |
| 4.62e-143 | 54 | 476 | 56 | 507 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.39e-126 | 37 | 476 | 35 | 445 | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei [Rhizomucor miehei],4LYP_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei [Rhizomucor miehei] |
|
| 6.72e-125 | 37 | 476 | 35 | 445 | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant [Rhizomucor miehei] |
|
| 6.72e-125 | 37 | 476 | 35 | 445 | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant [Rhizomucor miehei],4NRR_A Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose [Rhizomucor miehei],4NRR_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose [Rhizomucor miehei],4NRS_A Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose [Rhizomucor miehei],4NRS_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose [Rhizomucor miehei] |
|
| 5.99e-64 | 36 | 458 | 22 | 415 | Exo-mannosidase from Cellvibrio mixtus [Cellvibrio mixtus],1UZ4_A Common inhibition of beta-glucosidase and beta-mannosidase by isofagomine lactam reflects different conformational intineraries for glucoside and mannoside hydrolysis [Cellvibrio mixtus],7ODJ_AAA Chain AAA, Man5A [Cellvibrio mixtus] |
|
| 7.53e-34 | 36 | 428 | 4 | 335 | Chain A, endo-beta-mannanase [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.86e-34 | 23 | 428 | 18 | 361 | Mannan endo-1,4-beta-mannosidase 4 OS=Solanum lycopersicum OX=4081 GN=MAN4 PE=1 SV=2 |
|
| 9.46e-34 | 27 | 386 | 9 | 335 | Mannan endo-1,4-beta-mannosidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN4 PE=2 SV=2 |
|
| 2.92e-33 | 52 | 365 | 45 | 322 | Mannan endo-1,4-beta-mannosidase 7 OS=Arabidopsis thaliana OX=3702 GN=MAN7 PE=2 SV=1 |
|
| 3.74e-33 | 41 | 412 | 34 | 357 | Putative mannan endo-1,4-beta-mannosidase 9 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN9 PE=2 SV=2 |
|
| 3.82e-32 | 32 | 373 | 33 | 339 | Mannan endo-1,4-beta-mannosidase 2 OS=Solanum lycopersicum OX=4081 GN=MAN2 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
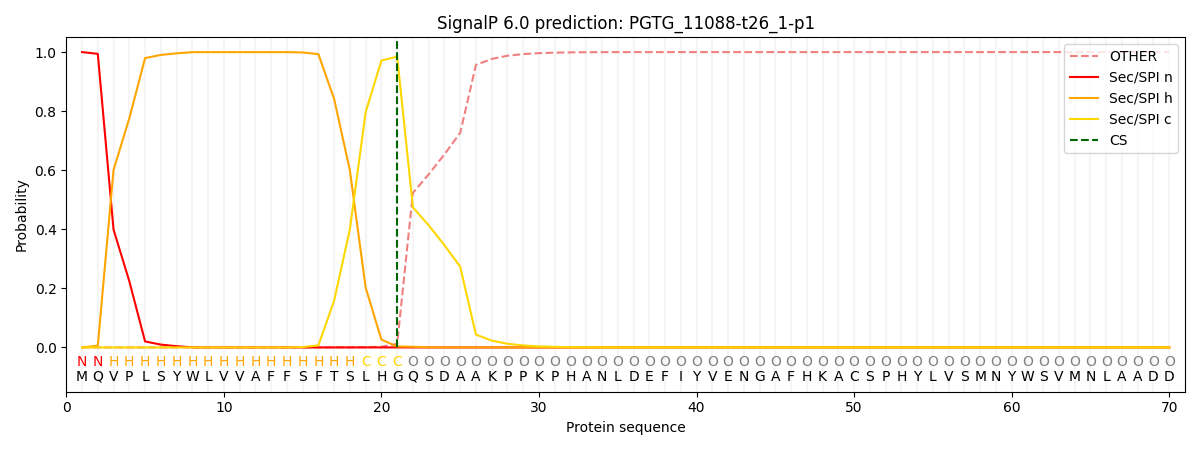
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000207 | 0.999775 | CS pos: 21-22. Pr: 0.9847 |
