You are browsing environment: FUNGIDB
CAZyme Information: PGTG_02189-t26_1-p1
You are here: Home > Sequence: PGTG_02189-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia graminis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia graminis | |||||||||||
| CAZyme ID | PGTG_02189-t26_1-p1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 236776 | PRK10856 | 2.72e-05 | 88 | 168 | 170 | 252 | cytoskeleton protein RodZ. |
| 165513 | PHA03255 | 3.47e-05 | 79 | 166 | 45 | 134 | BDLF3; Provisional |
| 411196 | act_recrut_TARP | 7.33e-05 | 77 | 271 | 357 | 543 | type III secretion system actin-recruiting effector Tarp. The founding member of the Tarp (translocated actin-recruiting phosphoprotein) is CT456 from Chlamydia trachomatis. Tarp is a type III secretion system effector. Orthologs are found other Chlamydia, but are highly variable in length because many lack much of the repeat region. |
| 282904 | Herpes_BLLF1 | 5.11e-04 | 89 | 166 | 491 | 570 | Herpes virus major outer envelope glycoprotein (BLLF1). This family consists of the BLLF1 viral late glycoprotein, also termed gp350/220. It is the most abundantly expressed glycoprotein in the viral envelope of the Herpesviruses and is the major antigen responsible for stimulating the production of neutralising antibodies in vivo. |
| 237019 | PRK11907 | 0.002 | 88 | 167 | 34 | 111 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.56e-130 | 178 | 459 | 180 | 458 | |
| 2.96e-68 | 180 | 438 | 15 | 285 | |
| 4.05e-67 | 180 | 438 | 14 | 284 | |
| 8.23e-67 | 180 | 438 | 14 | 284 | |
| 1.86e-64 | 180 | 438 | 12 | 282 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
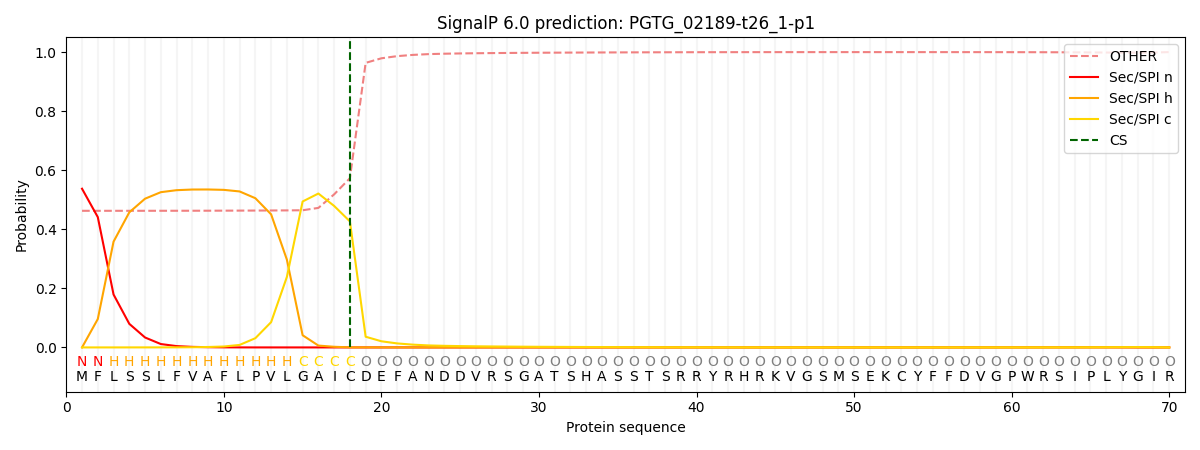
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.480831 | 0.519144 | CS pos: 18-19. Pr: 0.4268 |
