You are browsing environment: FUNGIDB
CAZyme Information: PGTG_01401-t26_1-p1
Basic Information
help
| Species |
Puccinia graminis
|
| Lineage |
Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia graminis
|
| CAZyme ID |
PGTG_01401-t26_1-p1
|
| CAZy Family |
AA7 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 570 |
|
61719.83 |
5.3470 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PgraminisCRL75-36-700-3 |
16309 |
418459 |
509 |
15800
|
|
| Gene Location |
No EC number prediction in PGTG_01401-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| PL35 |
302 |
490 |
7e-44 |
0.9832402234636871 |
PGTG_01401-t26_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.07e-200 |
31 |
570 |
248 |
790 |
| 2.43e-199 |
31 |
570 |
248 |
790 |
| 1.62e-196 |
36 |
569 |
171 |
696 |
| 2.67e-195 |
31 |
570 |
257 |
799 |
| 1.20e-192 |
34 |
570 |
158 |
696 |
PGTG_01401-t26_1-p1 has no PDB hit.
PGTG_01401-t26_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
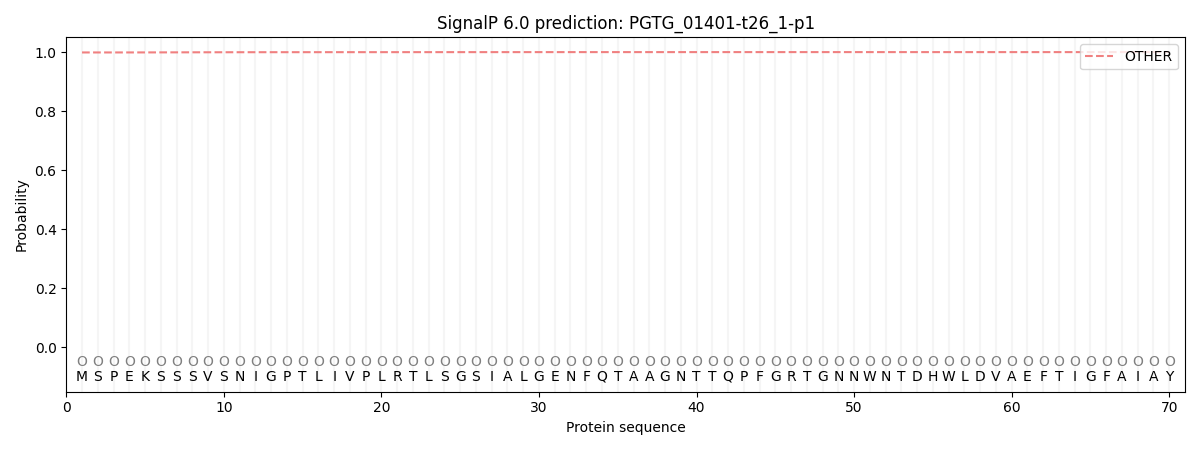
| Other |
SP_Sec_SPI |
CS Position |
| 0.998921 |
0.001083 |
|
There is no transmembrane helices in PGTG_01401-t26_1-p1.

