You are browsing environment: FUNGIDB
CAZyme Information: PGTG_01262-t26_1-p1
Basic Information
help
| Species |
Puccinia graminis
|
| Lineage |
Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia graminis
|
| CAZyme ID |
PGTG_01262-t26_1-p1
|
| CAZy Family |
GT90 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 996 |
|
111700.29 |
6.5153 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PgraminisCRL75-36-700-3 |
16309 |
418459 |
509 |
15800
|
|
| Gene Location |
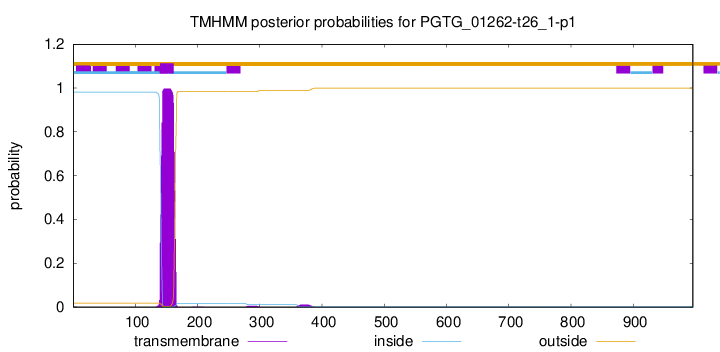
PGTG_01262-t26_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 9.66e-171 |
137 |
687 |
321 |
896 |
| 2.68e-170 |
137 |
687 |
321 |
896 |
| 2.98e-169 |
138 |
805 |
333 |
1018 |
| 3.60e-169 |
137 |
687 |
320 |
901 |
| 5.34e-169 |
137 |
687 |
327 |
897 |
PGTG_01262-t26_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.23e-11 |
195 |
446 |
63 |
327 |
Beta-1,3-galactosyltransferase pvg3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=pvg3 PE=1 SV=1 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
CS Position |
| 1.000023 |
0.000028 |
|