You are browsing environment: FUNGIDB
CAZyme Information: PAAG_12274-t30_1-p1
You are here: Home > Sequence: PAAG_12274-t30_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paracoccidioides lutzii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; NA; Paracoccidioides; Paracoccidioides lutzii | |||||||||||
| CAZyme ID | PAAG_12274-t30_1-p1 | |||||||||||
| CAZy Family | GT69 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 153 | 239 | 7.2e-20 | 0.5608465608465608 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 8.29e-33 | 101 | 239 | 2 | 166 | Cutinase. |
| 369775 | PE-PPE | 8.56e-04 | 141 | 203 | 31 | 106 | PE-PPE domain. This domain is found C terminal to the PE (pfam00934) and PPE (pfam00823) domains. The secondary structure of this domain is predicted to be a mixture of alpha helices and beta strands. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.84e-83 | 35 | 244 | 2 | 232 | |
| 1.12e-77 | 43 | 244 | 54 | 276 | |
| 1.46e-44 | 92 | 244 | 104 | 275 | |
| 1.67e-44 | 42 | 244 | 112 | 324 | |
| 7.21e-44 | 42 | 244 | 112 | 328 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.37e-26 | 97 | 242 | 75 | 244 | Structure of cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
|
| 9.46e-17 | 91 | 239 | 5 | 185 | Humicola insolens cutinase [Humicola insolens],4OYY_B Humicola insolens cutinase [Humicola insolens],4OYY_C Humicola insolens cutinase [Humicola insolens],4OYY_D Humicola insolens cutinase [Humicola insolens],4OYY_E Humicola insolens cutinase [Humicola insolens],4OYY_F Humicola insolens cutinase [Humicola insolens],4OYY_G Humicola insolens cutinase [Humicola insolens],4OYY_H Humicola insolens cutinase [Humicola insolens],4OYY_I Humicola insolens cutinase [Humicola insolens],4OYY_J Humicola insolens cutinase [Humicola insolens],4OYY_K Humicola insolens cutinase [Humicola insolens],4OYY_L Humicola insolens cutinase [Humicola insolens] |
|
| 9.33e-15 | 91 | 239 | 5 | 185 | Humicola insolens cutinase in complex with mono-ethylphosphate [Humicola insolens],4OYL_B Humicola insolens cutinase in complex with mono-ethylphosphate [Humicola insolens],4OYL_C Humicola insolens cutinase in complex with mono-ethylphosphate [Humicola insolens] |
|
| 6.46e-13 | 91 | 242 | 9 | 190 | Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY3_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY9_A Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9],7CY9_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9] |
|
| 1.71e-11 | 101 | 230 | 23 | 183 | Chain A, cutinase [Malbranchea cinnamomea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.27e-42 | 88 | 244 | 47 | 220 | Cutinase 4 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cut4 PE=2 SV=1 |
|
| 9.80e-28 | 97 | 241 | 75 | 243 | Cutinase cut1 OS=Trichoderma harzianum OX=5544 GN=cut1 PE=1 SV=1 |
|
| 3.83e-26 | 82 | 241 | 18 | 196 | Cutinase OS=Botryotinia fuckeliana OX=40559 GN=cutA PE=1 SV=1 |
|
| 1.53e-25 | 97 | 242 | 75 | 244 | Cutinase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=TRIREDRAFT_60489 PE=3 SV=2 |
|
| 1.53e-25 | 97 | 242 | 75 | 244 | Cutinase OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=M419DRAFT_76732 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
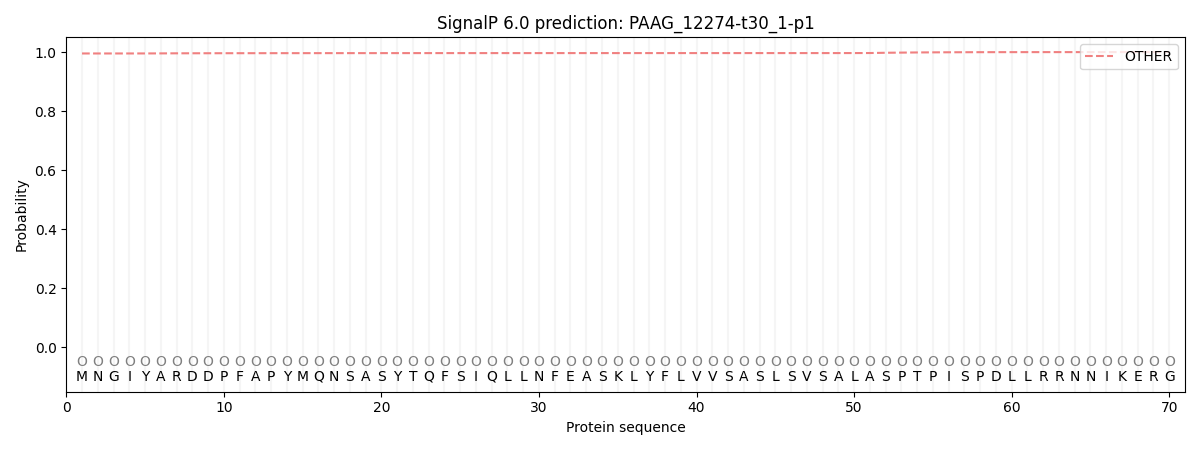
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.995789 | 0.004202 |
