You are browsing environment: FUNGIDB
CAZyme Information: PAAG_07832-t30_1-p1
You are here: Home > Sequence: PAAG_07832-t30_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
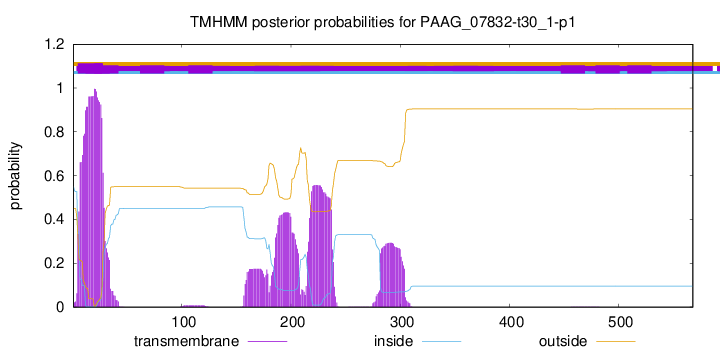
TMHMM annotations
Basic Information help
| Species | Paracoccidioides lutzii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; NA; Paracoccidioides; Paracoccidioides lutzii | |||||||||||
| CAZyme ID | PAAG_07832-t30_1-p1 | |||||||||||
| CAZy Family | GT32 | |||||||||||
| CAZyme Description | alpha-1,2-mannosyltransferase alg-11 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.131:1 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340835 | GT4_ALG11-like | 0.0 | 101 | 545 | 1 | 419 | alpha-1,2-mannosyltransferase ALG11 and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. ALG11 in yeast is involved in adding the final 1,2-linked Man to the Man5GlcNAc2-PP-Dol synthesized on the cytosolic face of the ER. The deletion analysis of ALG11 was shown to block the early steps of core biosynthesis that takes place on the cytoplasmic face of the ER and lead to a defect in the assembly of lipid-linked oligosaccharides. |
| 406369 | ALG11_N | 1.11e-122 | 101 | 308 | 1 | 208 | ALG11 mannosyltransferase N-terminus. |
| 215511 | PLN02949 | 5.06e-118 | 103 | 555 | 36 | 459 | transferase, transferring glycosyl groups |
| 395425 | Glycos_transf_1 | 1.75e-16 | 348 | 500 | 2 | 124 | Glycosyl transferases group 1. Mutations in this domain of PIGA lead to disease (Paroxysmal Nocturnal haemoglobinuria). Members of this family transfer activated sugars to a variety of substrates, including glycogen, Fructose-6-phosphate and lipopolysaccharides. Members of this family transfer UDP, ADP, GDP or CMP linked sugars. The eukaryotic glycogen synthases may be distant members of this family. |
| 223515 | RfaB | 2.98e-14 | 102 | 546 | 3 | 370 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 566 | 1 | 556 | |
| 0.0 | 5 | 566 | 4 | 555 | |
| 1.03e-317 | 1 | 566 | 1 | 556 | |
| 1.91e-248 | 11 | 557 | 10 | 551 | |
| 5.46e-248 | 7 | 559 | 2 | 546 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.76e-180 | 16 | 556 | 11 | 546 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-11 PE=3 SV=1 |
|
| 1.84e-113 | 103 | 554 | 57 | 464 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg11 PE=3 SV=1 |
|
| 2.33e-93 | 102 | 554 | 64 | 491 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Pongo abelii OX=9601 GN=ALG11 PE=2 SV=2 |
|
| 2.85e-93 | 102 | 542 | 62 | 474 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Xenopus tropicalis OX=8364 GN=alg11 PE=2 SV=1 |
|
| 7.71e-93 | 102 | 542 | 60 | 473 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Xenopus laevis OX=8355 GN=alg11 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
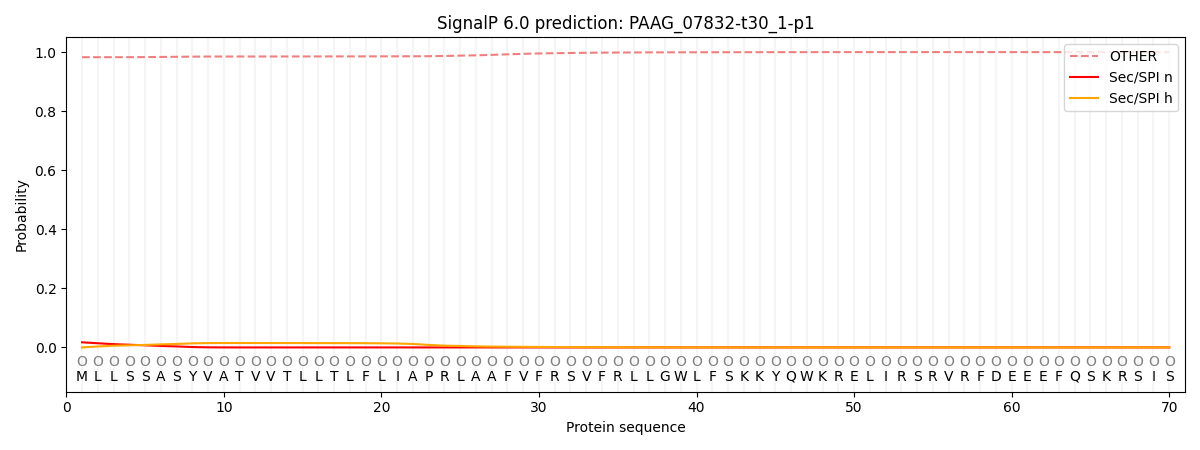
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.983812 | 0.016217 |