You are browsing environment: FUNGIDB
CAZyme Information: P175DRAFT_0509856-t39_1-p1
You are here: Home > Sequence: P175DRAFT_0509856-t39_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus ochraceoroseus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus ochraceoroseus | |||||||||||
| CAZyme ID | P175DRAFT_0509856-t39_1-p1 | |||||||||||
| CAZy Family | GH72|CBM43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT32 | 81 | 159 | 6.9e-19 | 0.9222222222222223 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226297 | OCH1 | 3.46e-26 | 40 | 262 | 60 | 247 | Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| 308657 | TPT | 1.15e-18 | 256 | 545 | 2 | 290 | Triose-phosphate Transporter family. This family includes transporters with a specificity for triose phosphate. |
| 411044 | TPT_S35C2 | 2.60e-16 | 321 | 544 | 2 | 234 | solute carrier family 35 member C2, member of the triose-phosphate transporter family. Solute carrier family 35 member C2 (S35C2 or Slc35c2), also called ovarian cancer-overexpressed gene 1 protein (OVCOV1), is a member of the triose-phosphate transporter (TPT) family, which is part of the drug/metabolite transporter (DMT) superfamily. It may function either as a GDP-fucose transporter that competes with Slc35c1 (S35C1) for GDP-fucose, or a factor that otherwise enhances the fucosylation of Notch and is required for optimal Notch signaling in mammalian cells. |
| 398274 | Gly_transf_sug | 3.30e-16 | 85 | 159 | 8 | 86 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
| 240371 | PTZ00343 | 3.83e-15 | 255 | 546 | 48 | 348 | triose or hexose phosphate/phosphate translocator; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.27e-164 | 3 | 552 | 79 | 739 | |
| 1.08e-102 | 6 | 238 | 12 | 246 | |
| 1.08e-102 | 6 | 238 | 12 | 246 | |
| 1.52e-102 | 6 | 238 | 12 | 246 | |
| 2.15e-102 | 6 | 238 | 12 | 246 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.03e-38 | 264 | 545 | 25 | 306 | Probable sugar phosphate/phosphate translocator At5g11230 OS=Arabidopsis thaliana OX=3702 GN=At5g11230 PE=2 SV=1 |
|
| 5.02e-38 | 264 | 545 | 25 | 306 | Probable sugar phosphate/phosphate translocator At4g32390 OS=Arabidopsis thaliana OX=3702 GN=At4g32390 PE=3 SV=1 |
|
| 1.77e-37 | 264 | 545 | 25 | 306 | Probable sugar phosphate/phosphate translocator At5g25400 OS=Arabidopsis thaliana OX=3702 GN=At5g25400 PE=2 SV=1 |
|
| 4.44e-37 | 264 | 545 | 25 | 306 | Probable sugar phosphate/phosphate translocator At2g25520 OS=Arabidopsis thaliana OX=3702 GN=At2g25520 PE=1 SV=1 |
|
| 6.19e-33 | 262 | 545 | 22 | 304 | Probable sugar phosphate/phosphate translocator At1g53660 OS=Arabidopsis thaliana OX=3702 GN=At1g53660 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000031 | 0.000010 |
TMHMM Annotations download full data without filtering help
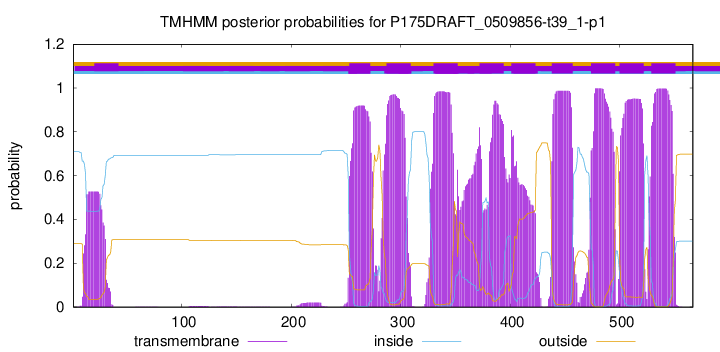
| Start | End |
|---|---|
| 253 | 272 |
| 287 | 309 |
| 330 | 352 |
| 372 | 394 |
| 401 | 423 |
| 438 | 457 |
| 474 | 496 |
| 500 | 522 |
| 529 | 551 |
