You are browsing environment: FUNGIDB
CAZyme Information: P175DRAFT_0472569-t39_1-p1
You are here: Home > Sequence: P175DRAFT_0472569-t39_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
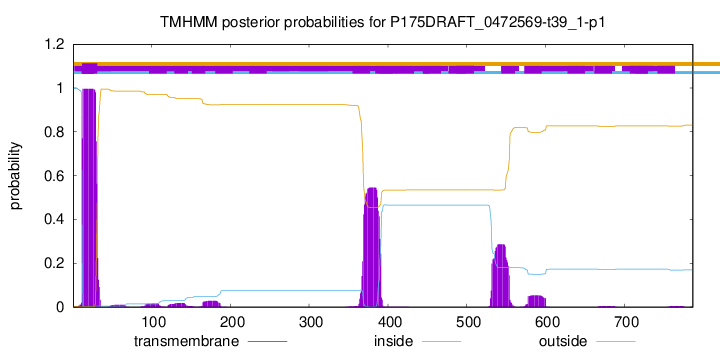
TMHMM annotations
Basic Information help
| Species | Aspergillus ochraceoroseus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus ochraceoroseus | |||||||||||
| CAZyme ID | P175DRAFT_0472569-t39_1-p1 | |||||||||||
| CAZy Family | GH154 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT34 | 66 | 257 | 5e-28 | 0.8861788617886179 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 410682 | CYP_fungal | 3.99e-111 | 435 | 787 | 1 | 343 | unknown subfamily of fungal cytochrome P450s. This subfamily is composed of uncharacterized fungal cytochrome P450s. Cytochrome P450 (P450, CYP) is a large superfamily of heme-containing proteins that catalyze a variety of oxidative reactions of a large number of structurally different endogenous and exogenous compounds in organisms from all major domains of life. CYPs bind their diverse ligands in a buried, hydrophobic active site, which is accessed through a substrate access channel formed by two flexible helices and their connecting loop. Their monooxygenase activity relies on the reductive scission of molecular oxygen bound to the P450 heme iron, and the delivery of two electrons to the heme iron during the catalytic cycle. |
| 410684 | CYP67-like | 5.81e-80 | 435 | 786 | 1 | 337 | cytochrome P450 family 67 and similar cytochrome P450s. This subfamily includes Uromyces viciae-fabae cytochrome P450 67 (CYP67), also called planta-induced rust protein 16, Cystobasidium minutum (Rhodotorula minuta) cytochrome P450rm, and other fungal cytochrome P450s. P450rm catalyzes the formation of isobutene and 4-hydroxylation of benzoate. The gene encoding CYP67 is a planta-induced gene that is expressed in haustoria and rust-infected leaves. The CYP67-like subfamily belongs to the large cytochrome P450 (P450, CYP) superfamily of heme-containing proteins that catalyze a variety of oxidative reactions of a large number of structurally different endogenous and exogenous compounds in organisms from all major domains of life. CYPs bind their diverse ligands in a buried, hydrophobic active site, which is accessed through a substrate access channel formed by two flexible helices and their connecting loop. |
| 410681 | CYP60B-like | 5.27e-71 | 435 | 787 | 1 | 338 | cytochrome P450 family 60, subfamily B and similar cytochrome P450s. This family is composed of fungal cytochrome P450s including: Aspergillus nidulans cytochrome P450 60B (CYP60B), also called versicolorin B desaturase, which catalyzes the conversion of versicolorin B to versicolorin A during sterigmatocystin biosynthesis; Fusarium sporotrichioides cytochrome P450 65A1 (CYP65A1), also called isotrichodermin C-15 hydroxylase, which catalyzes the hydroxylation at C-15 of isotricodermin in trichothecene biosynthesis; and Penicillium aethiopicum P450 monooxygenase vrtK, also called viridicatumtoxin synthesis protein K, which catalyzes the spirocyclization of the geranyl moiety of previridicatumtoxin to produce viridicatumtoxin, a tetracycline-like fungal meroterpenoid. The CYP60B-like family belongs to the large cytochrome P450 (P450, CYP) superfamily of heme-containing proteins that catalyze a variety of oxidative reactions of a large number of structurally different endogenous and exogenous compounds in organisms from all major domains of life. CYPs bind their diverse ligands in a buried, hydrophobic active site, which is accessed through a substrate access channel formed by two flexible helices and their connecting loop. |
| 410685 | CYP58-like | 5.68e-69 | 435 | 787 | 1 | 346 | cytochrome P450 family 58-like fungal cytochrome P450s. This group includes Fusarium sporotrichioides cytochrome P450 58 (CYP58, also known as Tri4 and trichodiene oxygenase), and similar fungal proteins. CYP58 catalyzes the oxygenation of trichodiene during the biosynthesis of trichothecenes, which are sesquiterpenoid toxins that act by inhibiting protein biosynthesis. The CYP58-like subfamily belongs to the large cytochrome P450 (P450, CYP) superfamily of heme-containing proteins that catalyze a variety of oxidative reactions of a large number of structurally different endogenous and exogenous compounds in organisms from all major domains of life. CYPs bind their diverse ligands in a buried, hydrophobic active site, which is accessed through a substrate access channel formed by two flexible helices and their connecting loop. |
| 410683 | CYP57A1-like | 1.56e-67 | 435 | 787 | 1 | 352 | cytochrome P450 family 57, subfamily A, polypeptide 1 and similar cytochrome P450s. This family is composed of fungal cytochrome P450s including: Nectria haematococca cytochrome P450 57A1 (CYP57A1), also called pisatin demethylase, which detoxifies the phytoalexin pisatin; Penicillium aethiopicum P450 monooxygenase gsfF, also called griseofulvin synthesis protein F, which catalyzes the coupling of orcinol and phloroglucinol rings in griseophenone B to form desmethyl-dehydrogriseofulvin A during the biosynthesis of griseofulvin, a spirocyclic fungal natural product used to treat dermatophyte infections; and Penicillium aethiopicum P450 monooxygenase vrtE, also called viridicatumtoxin synthesis protein E, which catalyzes hydroxylation at C5 of the polyketide backbone during the biosynthesis of viridicatumtoxin, a tetracycline-like fungal meroterpenoid. The CYP57A1-like family belongs to the large cytochrome P450 (P450, CYP) superfamily of heme-containing proteins that catalyze a variety of oxidative reactions of a large number of structurally different endogenous and exogenous compounds in organisms from all major domains of life. CYPs bind their diverse ligands in a buried, hydrophobic active site, which is accessed through a substrate access channel formed by two flexible helices and their connecting loop. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 7 | 787 | 5 | 790 | |
| 5.03e-144 | 8 | 354 | 10 | 357 | |
| 1.37e-125 | 6 | 349 | 3 | 349 | |
| 6.01e-124 | 51 | 351 | 28 | 326 | |
| 2.53e-121 | 51 | 351 | 45 | 342 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.37e-14 | 478 | 787 | 95 | 393 | Human CYP3A4 bound to an inhibitor fluconazole [Homo sapiens] |
|
| 1.81e-14 | 391 | 787 | 5 | 393 | Crystal structure of the cysteine depleted CYP3A4 bound to glycerol [Homo sapiens],5VCE_A Crystal structure of the cysteine depleted CYP3A4 bound to ritonavir [Homo sapiens],5VCG_A Crystal structure of the cysteine depleted CYP3A4 bound to bromoergocryptine [Homo sapiens] |
|
| 5.51e-14 | 478 | 787 | 92 | 390 | Crystal structure of CYP3A4 in complex with an inhibitor [Homo sapiens] |
|
| 5.53e-14 | 478 | 787 | 93 | 391 | Crystal structure of human cytochrome P450 3A4 [Homo sapiens],1W0F_A Crystal structure of human cytochrome P450 3A4 [Homo sapiens],1W0G_A Crystal structure of human cytochrome P450 3A4 [Homo sapiens],2J0D_A Crystal structure of human P450 3A4 in complex with erythromycin [Homo sapiens],2J0D_B Crystal structure of human P450 3A4 in complex with erythromycin [Homo sapiens],2V0M_A Crystal structure of human P450 3A4 in complex with ketoconazole [Homo sapiens],2V0M_B Crystal structure of human P450 3A4 in complex with ketoconazole [Homo sapiens],2V0M_C Crystal structure of human P450 3A4 in complex with ketoconazole [Homo sapiens],2V0M_D Crystal structure of human P450 3A4 in complex with ketoconazole [Homo sapiens],3NXU_A Crystal structure of human cytochrome P4503A4 bound to an inhibitor ritonavir [Homo sapiens],3NXU_B Crystal structure of human cytochrome P4503A4 bound to an inhibitor ritonavir [Homo sapiens] |
|
| 5.55e-14 | 478 | 787 | 94 | 392 | Crystal Structure of Human Microsomal P450 3A4 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.48e-98 | 388 | 787 | 25 | 405 | Cytochrome P450 monooxygenase orf2 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=orf2 PE=3 SV=1 |
|
| 1.02e-82 | 435 | 787 | 30 | 363 | Cytochrome P450 monooxygenase azaI OS=Aspergillus niger (strain ATCC 1015 / CBS 113.46 / FGSC A1144 / LSHB Ac4 / NCTC 3858a / NRRL 328 / USDA 3528.7) OX=380704 GN=azaI PE=2 SV=1 |
|
| 5.47e-57 | 394 | 786 | 56 | 430 | Cytochrome P450 monooxygenase stcB OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=stcB PE=2 SV=2 |
|
| 2.82e-54 | 379 | 786 | 30 | 421 | Cytochrome P450 monooxygenase cypX OS=Dothistroma septosporum (strain NZE10 / CBS 128990) OX=675120 GN=cypX PE=2 SV=1 |
|
| 2.82e-54 | 379 | 786 | 30 | 421 | Cytochrome P450 monooxygenase cypX OS=Dothistroma septosporum OX=64363 GN=cypX PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.966815 | 0.033209 |