You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_512137-t37_1-p1
You are here: Home > Sequence: P174DRAFT_512137-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
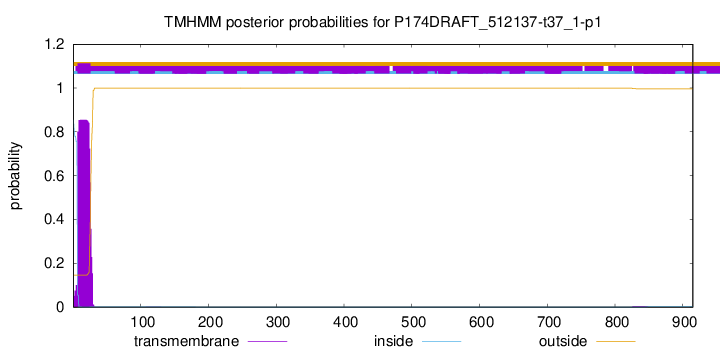
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_512137-t37_1-p1 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | exo-beta-1,3-glucanase Exg0 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.58:16 | 3.2.1.58:16 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 36 | 388 | 1.1e-143 | 0.4689189189189189 |
| GH55 | 532 | 912 | 2.2e-140 | 0.5581081081081081 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403800 | Pectate_lyase_3 | 1.63e-99 | 67 | 291 | 1 | 213 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 403800 | Pectate_lyase_3 | 4.31e-11 | 592 | 649 | 1 | 61 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 411474 | fibronec_FbpA | 5.78e-04 | 403 | 594 | 116 | 309 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
| 227721 | Pgu1 | 5.99e-04 | 69 | 119 | 84 | 125 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| 282904 | Herpes_BLLF1 | 0.001 | 416 | 574 | 398 | 565 | Herpes virus major outer envelope glycoprotein (BLLF1). This family consists of the BLLF1 viral late glycoprotein, also termed gp350/220. It is the most abundantly expressed glycoprotein in the viral envelope of the Herpesviruses and is the major antigen responsible for stimulating the production of neutralising antibodies in vivo. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 915 | 1 | 947 | |
| 0.0 | 1 | 915 | 1 | 950 | |
| 0.0 | 1 | 915 | 1 | 950 | |
| 0.0 | 1 | 915 | 1 | 950 | |
| 0.0 | 1 | 915 | 1 | 950 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.14e-269 | 43 | 915 | 4 | 756 | Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila],5M60_A Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila] |
|
| 9.99e-179 | 45 | 915 | 26 | 758 | Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQN_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_A Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium] |
|
| 2.47e-07 | 69 | 145 | 76 | 150 | Chain A, Putative pectin lyase [Geobacillus virus E2],7CHU_B Chain B, Putative pectin lyase [Geobacillus virus E2],7CHU_C Chain C, Putative pectin lyase [Geobacillus virus E2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 10 | 914 | 9 | 875 | Probable glucan endo-1,3-beta-glucosidase ARB_02077 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02077 PE=1 SV=1 |
|
| 1.71e-148 | 27 | 909 | 29 | 781 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
|
| 2.81e-35 | 565 | 899 | 359 | 744 | Glucan endo-1,3-beta-glucosidase BGN13.1 OS=Trichoderma harzianum OX=5544 GN=bgn13.1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
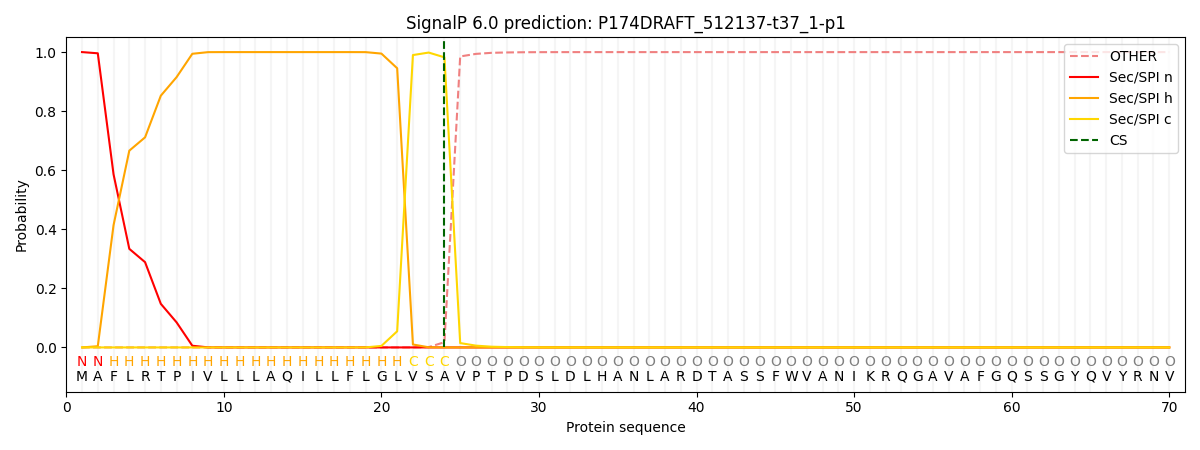
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000179 | 0.999814 | CS pos: 24-25. Pr: 0.9823 |