You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_510984-t37_1-p1
You are here: Home > Sequence: P174DRAFT_510984-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_510984-t37_1-p1 | |||||||||||
| CAZy Family | GT34 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH131 | 19 | 255 | 2.3e-100 | 0.9921568627450981 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 408085 | GH131_N | 1.54e-78 | 19 | 245 | 1 | 246 | Glycoside hydrolase 131 catalytic N-terminal domain. This is the N-terminal domain found in glycoside hydrolase family 131 (GH131A) protein observed in Coprinopsis cinerea. GH131A exhibits bifunctional exo-beta-1,3-/-1,6- and endo-beta-1,4 activity toward beta-glucan. This domain is catalytic in nature though the catalytic mechanism of C. cinerea GH131A is different from that of typical glycosidases that use a pair of carboxylic acid residues as the catalytic residues. In the case of GH131A, Glu98 and His218 may form a catalytic dyad and Glu98 may activate His218 during catalysis. |
| 236776 | PRK10856 | 3.20e-05 | 276 | 332 | 184 | 240 | cytoskeleton protein RodZ. |
| 139494 | PRK13335 | 5.75e-05 | 245 | 345 | 77 | 179 | superantigen-like protein SSL3; Reviewed. |
| 236776 | PRK10856 | 3.34e-04 | 276 | 347 | 174 | 244 | cytoskeleton protein RodZ. |
| 273167 | rad23 | 0.002 | 276 | 329 | 75 | 128 | UV excision repair protein Rad23. All proteins in this family for which functions are known are components of a multiprotein complex used for targeting nucleotide excision repair to specific parts of the genome. In humans, Rad23 complexes with the XPC protein. This family is based on the phylogenomic analysis of JA Eisen (1999, Ph.D. Thesis, Stanford University). [DNA metabolism, DNA replication, recombination, and repair] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.41e-158 | 1 | 264 | 1 | 262 | |
| 1.05e-157 | 1 | 263 | 1 | 261 | |
| 1.05e-157 | 1 | 263 | 1 | 261 | |
| 1.05e-157 | 1 | 263 | 1 | 261 | |
| 6.05e-157 | 1 | 259 | 1 | 257 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.73e-101 | 17 | 257 | 1 | 244 | Chain A, Beta-glucanase [Podospora anserina],4LE3_B Chain B, Beta-glucanase [Podospora anserina],4LE3_C Chain C, Beta-glucanase [Podospora anserina],4LE3_D Chain D, Beta-glucanase [Podospora anserina],4LE4_A Chain A, Beta-glucanase [Podospora anserina],4LE4_B Chain B, Beta-glucanase [Podospora anserina],4LE4_C Chain C, Beta-glucanase [Podospora anserina],4LE4_D Chain D, Beta-glucanase [Podospora anserina] |
|
| 1.86e-97 | 17 | 259 | 1 | 237 | Chain A, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_B Chain B, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_C Chain C, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130],3W9A_D Chain D, Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea [Coprinopsis cinerea okayama7#130] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
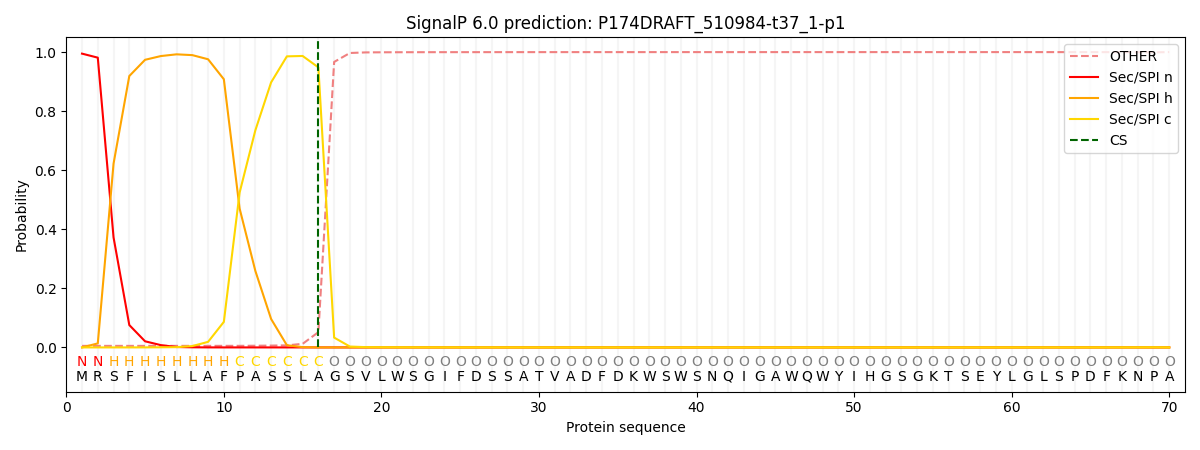
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.009721 | 0.990268 | CS pos: 16-17. Pr: 0.9482 |
