You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_464341-t37_1-p1
You are here: Home > Sequence: P174DRAFT_464341-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
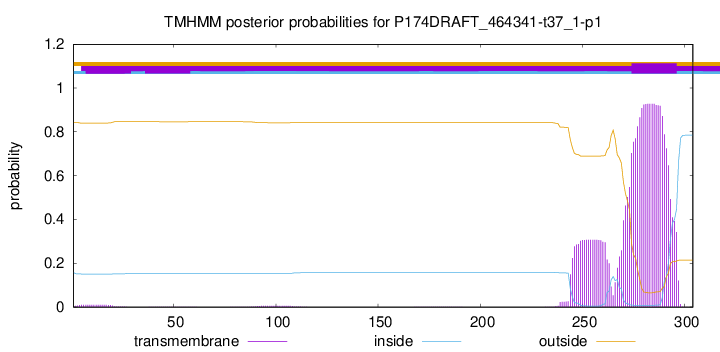
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_464341-t37_1-p1 | |||||||||||
| CAZy Family | GH78 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:67 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 42 | 206 | 9.9e-64 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 2.92e-100 | 41 | 204 | 1 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.55e-179 | 7 | 298 | 9 | 313 | |
| 1.81e-117 | 8 | 214 | 10 | 229 | |
| 2.46e-109 | 9 | 205 | 11 | 220 | |
| 2.46e-109 | 9 | 205 | 11 | 220 | |
| 2.46e-109 | 9 | 205 | 11 | 220 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.04e-90 | 30 | 208 | 1 | 190 | Crystal structure of family 11 xylanase in complex with inhibitor (XIP-I) [Talaromyces funiculosus] |
|
| 2.56e-90 | 25 | 208 | 13 | 207 | Xylanase 11C from Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) [Talaromyces funiculosus],3WP3_B Xylanase 11C from Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) [Talaromyces funiculosus] |
|
| 1.10e-87 | 32 | 208 | 3 | 190 | Chain A, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_B Chain B, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_C Chain C, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_D Chain D, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_E Chain E, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_F Chain F, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_G Chain G, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_H Chain H, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_I Chain I, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_J Chain J, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_K Chain K, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_L Chain L, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612] |
|
| 2.42e-78 | 24 | 211 | 18 | 218 | Crystal structure of environmental isolated GH11 in complex with xylobiose and feruloyl-arabino-xylotriose [Escherichia coli] |
|
| 2.50e-78 | 24 | 211 | 18 | 218 | Environmentally isolated GH11 xylanase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.37e-110 | 9 | 205 | 11 | 220 | Endo-1,4-beta-xylanase B OS=Penicillium oxalicum OX=69781 GN=xynB PE=1 SV=1 |
|
| 4.04e-98 | 25 | 208 | 22 | 216 | Endo-1,4-beta-xylanase 2 OS=Rhizopus oryzae OX=64495 GN=xyn2 PE=1 SV=1 |
|
| 3.43e-91 | 25 | 208 | 29 | 223 | Endo-1,4-beta-xylanase C OS=Talaromyces funiculosus OX=28572 GN=xynC PE=1 SV=1 |
|
| 4.54e-87 | 4 | 207 | 7 | 224 | Endo-1,4-beta-xylanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xlnB PE=3 SV=2 |
|
| 6.47e-86 | 15 | 208 | 16 | 221 | Endo-1,4-beta-xylanase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xlnB PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
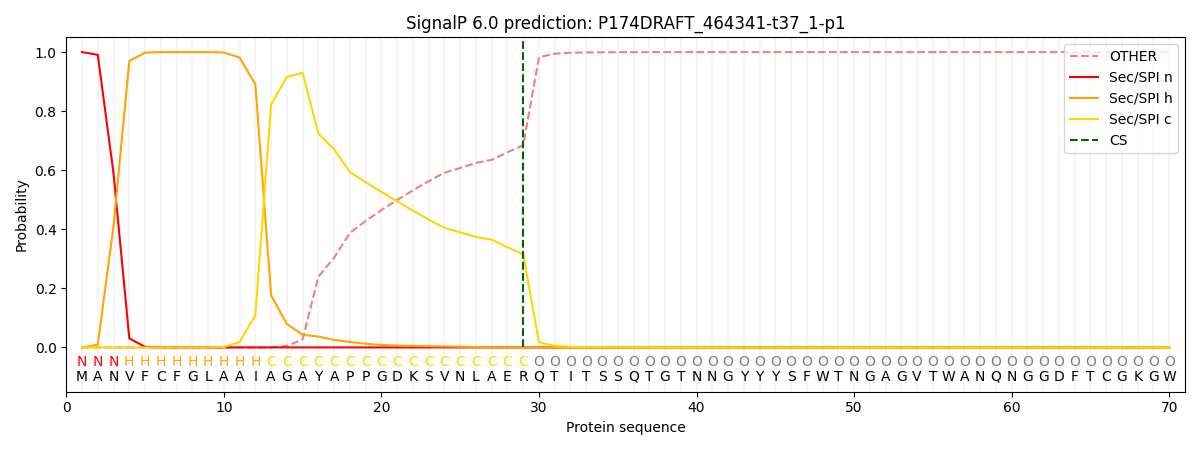
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000378 | 0.999624 | CS pos: 29-30. Pr: 0.3152 |