You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_463658-t37_1-p1
You are here: Home > Sequence: P174DRAFT_463658-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_463658-t37_1-p1 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | putative oligoxyloglucan-reducing end-specific xyloglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 806115; End:808562 Strand: + | |||||||||||
Full Sequence Download help
| MKYWLQQLGL TVLCALSAAA RTAEHAYEFK SVAITGGGYI TGIVGHPTEK NLLYARTDIG | 60 |
| STYRWEQGLN KWIPLTDFLG PEDENLLGTE SVAMDPTDPN RLYLAQGRYL SSNNSAFFVS | 120 |
| DDRGATFTRY RAPFPMGANE LGRNNGERLA VNPFKPNELW MGTRNAGLMR SSDRAKTWTN | 180 |
| VTNFPDAAAN GIGITFVIFD PKHEGAIYVG ASIPGGLYYT TDGGENWKSL PGQPMQWDPS | 240 |
| LLVYPNETQP QSAGPQPMKA VLASNGVLYV TYADYPGPWG VAYGAVHVYN TTASTWMDIT | 300 |
| PNANNTSPKP YTPQAFPAGG YCGLSVAADD PDTVVVVSLD RDPGPALDSM YLSRDGGKSW | 360 |
| KDVSQLSTPP GSGGYWGHPI AEAALANGTK VPWLSFNWGP QWGGYGAPSP VKGLTKVGWW | 420 |
| MTAVLIDPSN PDHVLYGTGA TIWATDTIAQ ADKNSTPKWY IQAQGIEETV TLAMISPREG | 480 |
| AHLLSGAGDI NGFRHDDLDT PQPMFGLPVF SNLNTLDWAG QRPEVIVRGG PCGHQYPDGC | 540 |
| GQAAYSTDGG SEWTKFQTCI KGVNTSSTNP GVMAIDASGK YVVWTSAMYV VSPSVQAVTP | 600 |
| AANDSGPYAS NDWGKTWASP RGLTVQTPYI SADRVQPKTF YAFSGGVWYV STDGGLSYDA | 660 |
| FNATKLGLPA HTGAVPVVSV DRAGEIWLAL GSNGVYHTTD FGKRWKRITH KGTVADLITV | 720 |
| GAATHGSKKP ALFIRGSQGD PKKSNYGIYR SDDNGSTWDR VDDDTHRYGG FNLIQGDPRV | 780 |
| YGRVYLGTGG RGLLYADIVP ERSDKEGNVL GTGGI | 815 |
Enzyme Prediction help
| EC | 3.2.1.151:27 | 3.2.1.150:4 | 3.2.1.151:5 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH74 | 82 | 189 | 2.5e-18 | 0.463519313304721 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 271234 | Sialidase_non-viral | 3.88e-09 | 545 | 758 | 51 | 293 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| 406353 | Sortilin-Vps10 | 3.91e-06 | 115 | 230 | 1 | 112 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| 406353 | Sortilin-Vps10 | 6.18e-06 | 72 | 230 | 13 | 164 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| 406353 | Sortilin-Vps10 | 9.37e-06 | 608 | 764 | 4 | 167 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| 405545 | PSII_BNR | 8.23e-05 | 18 | 228 | 55 | 268 | Photosynthesis system II assembly factor YCF48. YCF48 is one of several assembly factors of the photosynthesis system II. The photosynthesis system II occurs in Cyanobacteria that are Gram-negative bacteria performing oxygenic photosynthesis. One of the three membranes surrounding these bacteria is the inner thylakoid membrane (TM) system that is localized within the cell and houses the large pigment-protein complexes of the photosynthetic electron transfer chain, i.e. Photosystem (PS) II, PSI, the cytochrome b6f complex, and the ATP synthase. YCF48 is necessary for efficient assembly and repair of the PSII. YCF48 is found predominantly in the thykaloid membrane. It is a BNR repeat protein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBG67009.1|GH74 | 0.0 | 1 | 815 | 1 | 815 |
| EAA64249.1|GH74|3.2.1.150 | 0.0 | 18 | 798 | 26 | 808 |
| BCS21231.1|GH74 | 0.0 | 8 | 798 | 8 | 796 |
| BAC22065.1|GH74|3.2.1.150 | 7.39e-316 | 9 | 806 | 10 | 793 |
| CAF02212.1|GH74|3.2.1.150 | 7.39e-316 | 9 | 806 | 10 | 793 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1SQJ_A | 4.26e-315 | 24 | 806 | 2 | 770 | Crystal Structure Analysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) [Geotrichum sp. M128],1SQJ_B Crystal Structure Analysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) [Geotrichum sp. M128] |
| 2EBS_A | 2.44e-314 | 24 | 806 | 2 | 770 | Chain A, Oligoxyloglucan reducing end-specific cellobiohydrolase [Geotrichum sp. M128],2EBS_B Chain B, Oligoxyloglucan reducing end-specific cellobiohydrolase [Geotrichum sp. M128] |
| 3A0F_A | 2.72e-240 | 28 | 796 | 11 | 758 | The crystal structure of Geotrichum sp. M128 xyloglucanase [Geotrichum sp. M128] |
| 4LGN_A | 2.01e-133 | 27 | 806 | 7 | 743 | The structure of Acidothermus cellulolyticus family 74 glycoside hydrolase [Acidothermus cellulolyticus 11B] |
| 7KN8_A | 6.02e-133 | 27 | 797 | 10 | 713 | Chain A, Cellulase [Xanthomonas campestris pv. campestris str. ATCC 33913],7KN8_B Chain B, Cellulase [Xanthomonas campestris pv. campestris str. ATCC 33913] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|A1DAU0|XGCA_NEOFI | 0.0 | 1 | 815 | 1 | 815 | Probable oligoxyloglucan-reducing end-specific xyloglucanase OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xgcA PE=3 SV=1 |
| sp|Q5BD38|XGCA_EMENI | 0.0 | 18 | 798 | 26 | 808 | Oligoxyloglucan-reducing end-specific xyloglucanase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgcA PE=1 SV=1 |
| sp|Q8J0D2|CBHRE_GEOS1 | 1.31e-316 | 9 | 806 | 10 | 793 | Oligoxyloglucan reducing end-specific cellobiohydrolase OS=Geotrichum sp. (strain M128) OX=203496 PE=1 SV=1 |
| sp|Q3MUH7|XG74_PAESP | 8.05e-146 | 2 | 815 | 8 | 783 | Xyloglucanase OS=Paenibacillus sp. OX=58172 GN=xeg74 PE=1 SV=1 |
| sp|A3DFA0|XG74_ACET2 | 1.46e-123 | 15 | 797 | 27 | 757 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xghA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
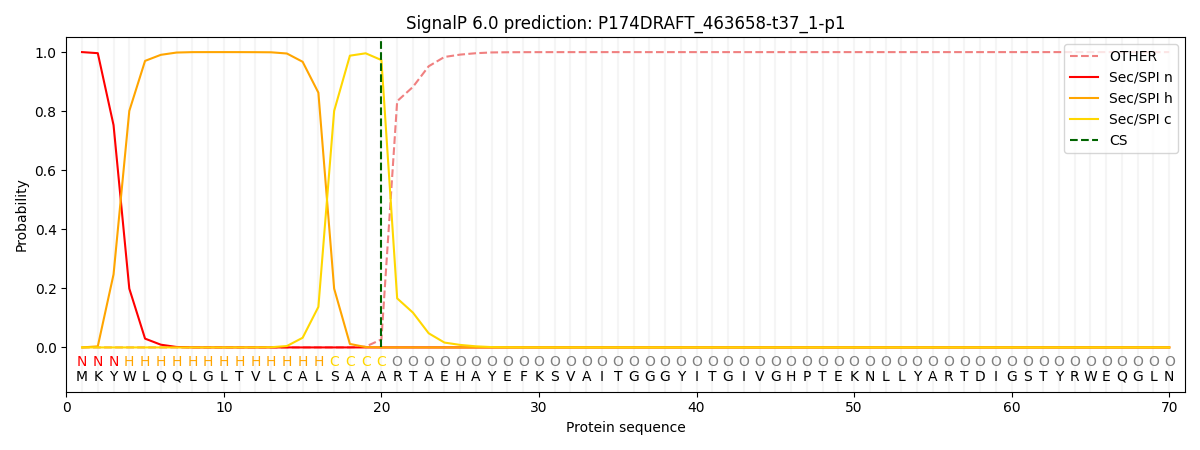
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000357 | 0.999620 | CS pos: 20-21. Pr: 0.9730 |
