You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_457101-t37_1-p1
You are here: Home > Sequence: P174DRAFT_457101-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_457101-t37_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | pectin lyase-like protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 68 | 386 | 9.4e-49 | 0.9076923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395231 | Glyco_hydro_28 | 1.28e-21 | 148 | 379 | 61 | 303 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| 215426 | PLN02793 | 5.18e-17 | 176 | 351 | 187 | 369 | Probable polygalacturonase |
| 177865 | PLN02218 | 6.91e-17 | 40 | 352 | 73 | 384 | polygalacturonase ADPG |
| 215540 | PLN03010 | 5.79e-16 | 176 | 401 | 167 | 396 | polygalacturonase |
| 227721 | Pgu1 | 9.12e-16 | 19 | 307 | 68 | 390 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.45e-294 | 13 | 478 | 12 | 487 | |
| 9.45e-294 | 13 | 478 | 12 | 487 | |
| 2.53e-287 | 6 | 478 | 6 | 489 | |
| 8.29e-269 | 13 | 466 | 12 | 475 | |
| 3.37e-268 | 13 | 466 | 12 | 475 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.49e-86 | 17 | 471 | 1 | 418 | Rhamnogalacturonase A From Aspergillus Aculeatus [Aspergillus aculeatus] |
|
| 1.28e-08 | 44 | 373 | 20 | 366 | Chain A, POLYGALACTURONASE [Pectobacterium carotovorum] |
|
| 2.09e-07 | 206 | 365 | 173 | 331 | Endo-Polygalacturonase Ii From Aspergillus Niger [Aspergillus niger],1CZF_B Endo-Polygalacturonase Ii From Aspergillus Niger [Aspergillus niger] |
|
| 1.05e-06 | 116 | 335 | 41 | 273 | Endopolygalacturonase I from Stereum purpureum at 0.96 A resolution [Chondrostereum purpureum],1KCC_A Endopolygalacturonase I from Stereum purpureum complexed with a galacturonate at 1.00 A resolution. [Chondrostereum purpureum],1KCD_A Endopolygalacturonase I from Stereum purpureum complexed with two galacturonate at 1.15 A resolution. [Chondrostereum purpureum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.54e-92 | 6 | 471 | 7 | 436 | Rhamnogalacturonase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=rhgA PE=2 SV=2 |
|
| 1.66e-91 | 3 | 466 | 6 | 432 | Probable rhamnogalacturonase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=rhgA PE=3 SV=1 |
|
| 4.66e-91 | 3 | 466 | 6 | 432 | Rhamnogalacturonase A OS=Aspergillus niger OX=5061 GN=rhgA PE=1 SV=1 |
|
| 7.62e-90 | 3 | 471 | 8 | 441 | Probable rhamnogalacturonase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=rhgB PE=3 SV=1 |
|
| 1.86e-89 | 3 | 471 | 8 | 441 | Probable rhamnogalacturonase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=rhgB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
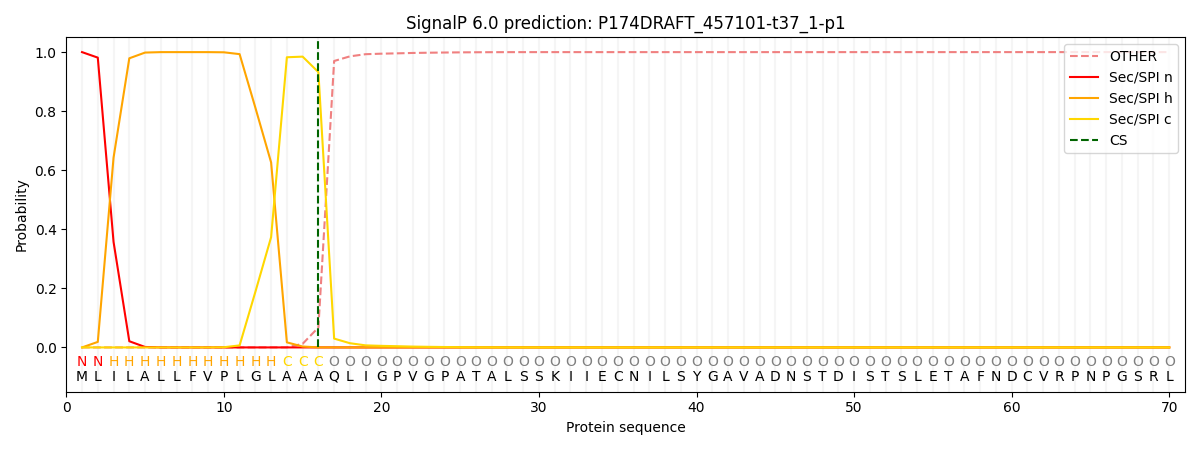
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000210 | 0.999736 | CS pos: 16-17. Pr: 0.9322 |
