You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_451754-t37_1-p1
You are here: Home > Sequence: P174DRAFT_451754-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_451754-t37_1-p1 | |||||||||||
| CAZy Family | PL26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215605 | PLN03153 | 1.48e-15 | 159 | 357 | 167 | 341 | hypothetical protein; Provisional |
| 398362 | DUF604 | 7.08e-09 | 277 | 357 | 6 | 90 | Protein of unknown function, DUF604. This family includes a conserved region found in several uncharacterized plant proteins. |
| 197223 | PLDc_like_TrmB_middle | 0.008 | 175 | 255 | 25 | 106 | Middle phospholipase D-like domain of the transcriptional regulator TrmB and similar proteins. Middle phospholipase D (PLD)-like domain of the transcriptional regulator TrmB and similar proteins. TrmB acts as a bifunctional sugar-sensing transcriptional regulator which controls two operons encoding maltose/trehalose and maltodextrin ABC transporters of Pyrococcus fruiosus. It functions as a dimer. Full length TrmB includes an N-terminal DNA-binding domain, a C-terminal sugar-binding domain and middle region that has been named as a PLD-like domain. The middle domain displays homology to PLD enzymes, which contain one or two HKD motifs (H-x-K-x(4)-D, where x represents any amino acid residue) per chain. The HKD motif characterizes the PLD superfamily. Due to the lack of key residues related to PLD activity in the PLD-like domain, members of this subfamily are unlikely to carry PLD activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 481 | 1 | 481 | |
| 1.33e-185 | 21 | 480 | 21 | 477 | |
| 1.33e-185 | 21 | 480 | 21 | 477 | |
| 6.17e-185 | 20 | 476 | 18 | 467 | |
| 8.75e-185 | 20 | 476 | 18 | 467 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
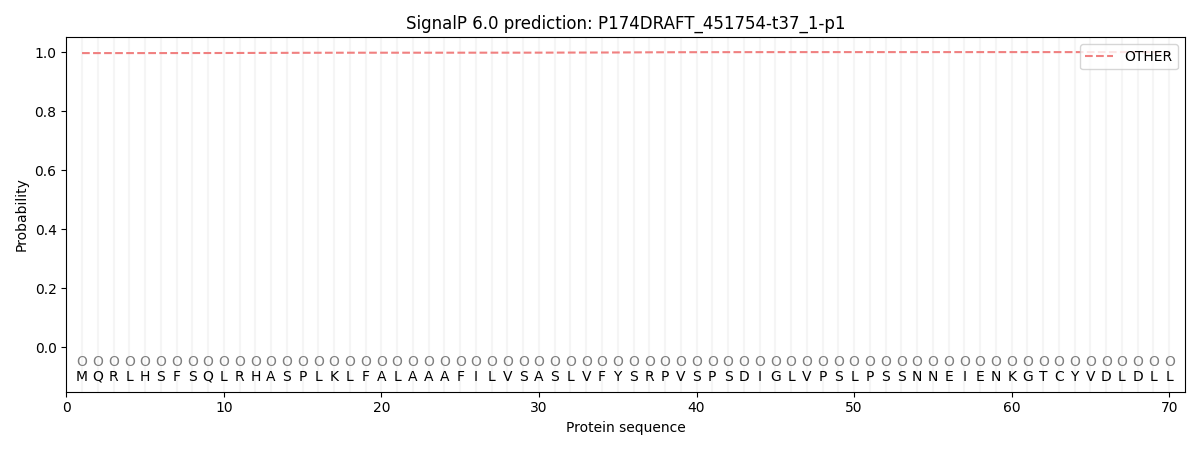
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.996880 | 0.003127 |

