You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_437520-t37_1-p1
You are here: Home > Sequence: P174DRAFT_437520-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_437520-t37_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 140 | 374 | 1.6e-47 | 0.9896907216494846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238871 | XynB_like | 1.05e-38 | 226 | 375 | 17 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 404371 | Lipase_GDSL_2 | 1.36e-12 | 213 | 363 | 6 | 173 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 238020 | FN3 | 3.37e-11 | 593 | 677 | 1 | 89 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| 238141 | SGNH_hydrolase | 2.16e-09 | 218 | 373 | 32 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 214495 | FN3 | 2.63e-08 | 593 | 671 | 1 | 83 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.49e-137 | 140 | 680 | 512 | 1051 | |
| 1.40e-123 | 139 | 679 | 335 | 864 | |
| 2.86e-65 | 142 | 589 | 345 | 773 | |
| 2.66e-59 | 139 | 391 | 138 | 383 | |
| 3.65e-55 | 137 | 393 | 54 | 309 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.84e-09 | 592 | 675 | 13 | 95 | Crystal structure of FN3con [synthetic construct] |
|
| 9.69e-06 | 589 | 677 | 4 | 93 | Chain A, Receptor-type tyrosine-protein phosphatase F [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.30e-06 | 357 | 648 | 385 | 663 | Collagen alpha-1(VII) chain OS=Homo sapiens OX=9606 GN=COL7A1 PE=1 SV=2 |
|
| 3.95e-06 | 593 | 643 | 170 | 220 | Exochitinase 1 OS=Streptomyces olivaceoviridis OX=1921 GN=chi01 PE=1 SV=1 |
|
| 9.35e-06 | 481 | 654 | 309 | 481 | Receptor-type tyrosine-protein phosphatase F OS=Homo sapiens OX=9606 GN=PTPRF PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
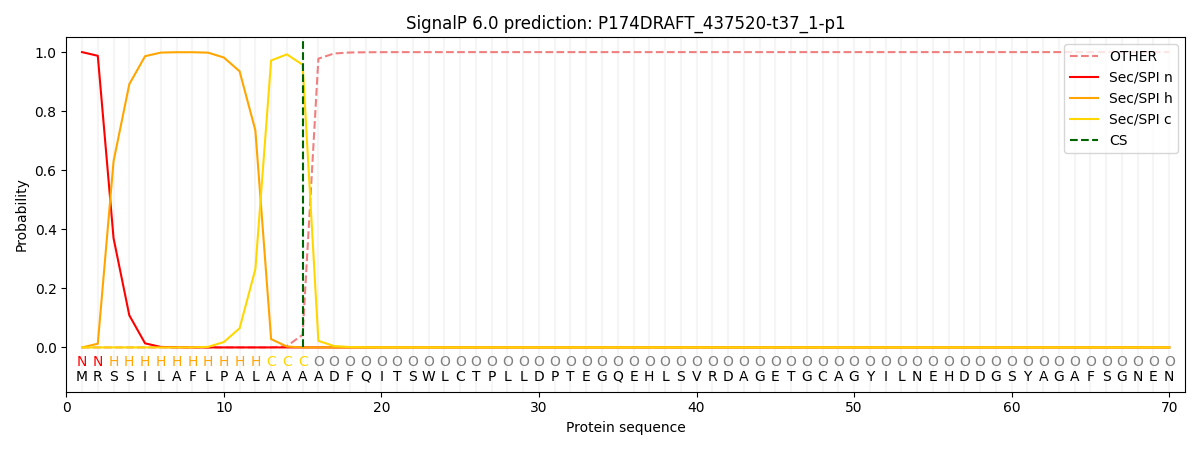
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000639 | 0.999334 | CS pos: 15-16. Pr: 0.9572 |

